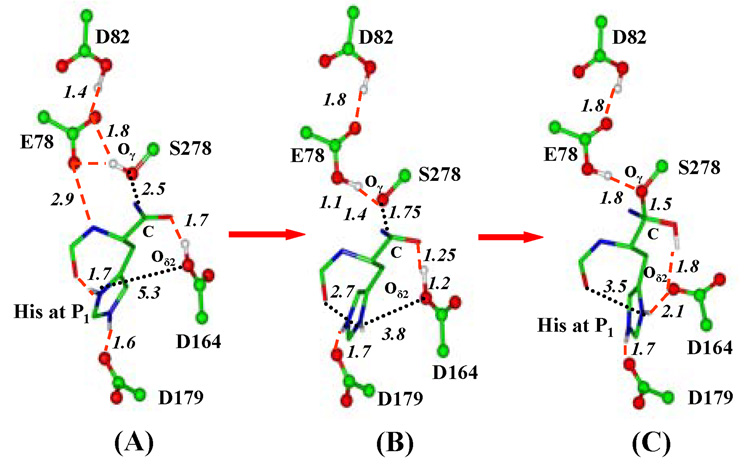
Figure 1.
Average structures of kumamolisin-As complex during the nucleophilic attack by Ser278 on substrate RPH*FR obtained from the QM/MM MD and free energy simulations. The enzyme is plotted in ball and stick and the ligand in stick. Hydrogen bonds (H-bonds) are indicated by red dashed lines. Ser278-Glu78-Asp82 is the catalytic triad. Only the sidechain of the P1 (His) residue for the substrate is shown for clarity. (A) The substrate complex. His sidechain at P1 donates an H-bond to the C=O group of Pro at P2, in addition to the carboxylate of Asp179. Glu78 accepts an H-bond from the backbone N-H group of the P1 residue (the H-bond distance shown here is between the non-hydrogen atoms). (B) An average structure near the transition state of the nucleophilic attack. The interaction of the His sidechain with the C=O group is significantly weakened and the H-bond between Glu78 and the N-H group is broken. (C) The TI complex. His at P1 has undergone a conformational change to interact with the unprotonated Asp164 residue and the H-bond to the C=O group observed in the substrate complex is broken.