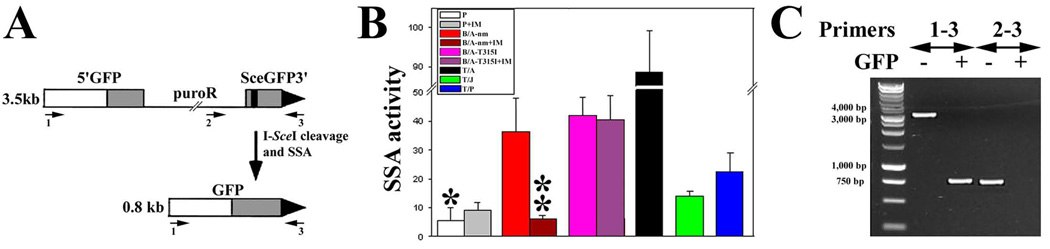
Figure 2. FTKs stimulate SSA.
(A) The structure of SA-GFP reporter cassette is shown before (upper panel, GFP− cells) and after (lower panel, GFP+ cells) I-SceI cleavage and SSA. The cassette consists of the 5’GFP and SceGFP3’ fragments, which have 266 bp of homology and intervening sequence encoding puromycin-resistance (puroR). The black strip represents the I-SceI site in the SceGFP3’ and the large black triangle depicts the 3’ end of the cassette. Repair of the I-SceI generated DSB in SceGFP3’ by SSA results in a functional GFP gene when a DNA strand from SceGFP3’ is annealed to the complementary strand of 5’GFP, followed by appropriate DNA-processing steps. As a result, SSA between the homologous sequences in the GFP gene fragments produces a 2.7-kb deletion in the chromosome. The SA-GFP reporter can also be repaired by HR and NHEJ, but without restoration of a functional GFP gene (14). (B) I-SceI and Red1-Mito were expressed in parental (P) and FTK-transformed (BCR/ABL non-mutated = B/A-nm, BCR/ABL-T315I mutant = B/A-T315I, TEL/ABL = T/A, TEL/JAK2 = T/J, and TEL/PDGFβR = T/P) cells containing SA-GFP reporter cassette and cultured in the presence of IL3 and imatinib (IM) when indicated. SSA activity was determined as the number of GFP+/Red1+ cells in 105 Red1+ cells. * p<10−8, <10−8, <10−8, 10−2 and <10−7 in comparison to B/A-nm, B/A-T315I, T/A, T/J and T/P, respectively; and ** p<0.03 in comparison to B/A. (C) PCR products from genomic DNA of GFP+ and GFP− cells.