Abstract
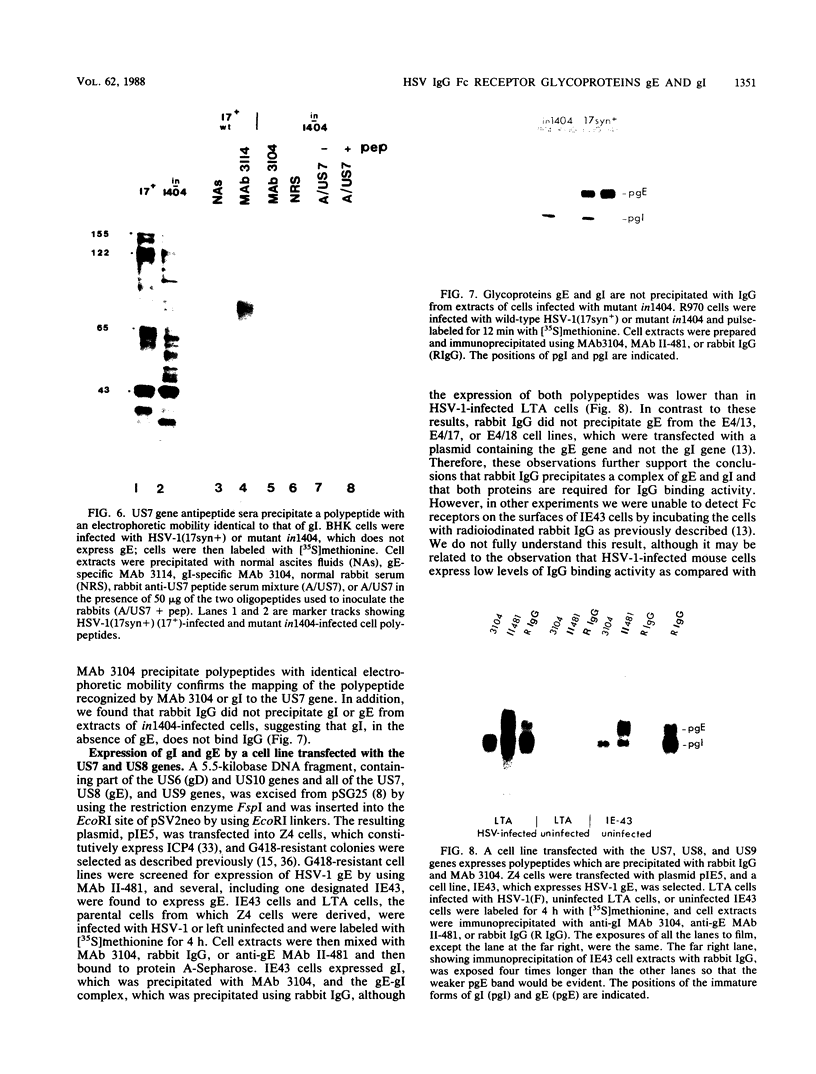
Evidence was recently presented that herpes simplex virus type 1 (HSV-1) immunoglobulin G (IgG) Fc receptors are composed of a complex containing a previously described glycoprotein, gE, and a novel virus-induced polypeptide, provisionally named g70 (D. C. Johnson and V. Feenstra, J. Virol. 61:2208-2216, 1987). Using a monoclonal antibody designated 3104, which recognizes g70, in conjunction with antipeptide sera and virus mutants unable to express g70 or gE, we have mapped the gene encoding g70 to the US7 open reading frame of HSV-1 adjacent to the gE gene. Therefore, g70 appears to be identical to a recently described polypeptide which was named gI (R. Longnecker, S. Chatterjee, R. J. Whitley, and B. Roizman, Proc. Natl. Acad. Sci. USA 84:147-151, 1987). Under mildly denaturing conditions, monoclonal antibody 3104 precipitated both gI and gE from extracts of HSV-1-infected cells. In addition, rabbit IgG precipitated the gE-gI complex from extracts of cells transfected with a fragment of HSV-1 DNA containing the gI, gE, and US9 genes. Cells infected with mutant viruses which were unable to express gE or gI did not bind radiolabeled IgG; however, cells coinfected with two viruses, one unable to express gE and the other unable to express gI, bound levels of IgG approaching those observed with wild-type viruses. These results further support the hypothesis that gE and gI form a complex which binds IgG by the Fc domain and that neither polypeptide alone can bind IgG.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler R., Glorioso J. C., Cossman J., Levine M. Possible role of Fc receptors on cells infected and transformed by herpesvirus: escape from immune cytolysis. Infect Immun. 1978 Aug;21(2):442–447. doi: 10.1128/iai.21.2.442-447.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucke R. B., Spear P. G. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J Virol. 1979 Dec;32(3):779–789. doi: 10.1128/jvi.32.3.779-789.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brown S. M., Ritchie D. A., Subak-Sharpe J. H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973 Mar;18(3):329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- Costa J., Rabson A. S., Yee C., Tralka T. S. Immunoglobulin binding to herpes virus-induced Fc receptors inhibits virus growth. Nature. 1977 Sep 15;269(5625):251–252. doi: 10.1038/269251a0. [DOI] [PubMed] [Google Scholar]
- Cross A. M., Hope R. G., Marsden H. S. Generation and properties of the glycoprotein E-related 32K/34K/35K and 55K/57K polypeptides encoded by herpes simplex virus type 1. J Gen Virol. 1987 Aug;68(Pt 8):2093–2104. doi: 10.1099/0022-1317-68-8-2093. [DOI] [PubMed] [Google Scholar]
- Frame M. C., McGeoch D. J., Rixon F. J., Orr A. C., Marsden H. S. The 10K virion phosphoprotein encoded by gene US9 from herpes simplex virus type 1. Virology. 1986 Apr 30;150(2):321–332. doi: 10.1016/0042-6822(86)90297-7. [DOI] [PubMed] [Google Scholar]
- Goldin A. L., Sandri-Goldin R. M., Levine M., Glorioso J. C. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J Virol. 1981 Apr;38(1):50–58. doi: 10.1128/jvi.38.1.50-58.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Heine J. W., Honess R. W., Cassai E., Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974 Sep;14(3):640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope R. G., Palfreyman J., Suh M., Marsden H. S. Sulphated glycoproteins induced by herpes simplex virus. J Gen Virol. 1982 Feb;58(Pt 2):399–415. doi: 10.1099/0022-1317-58-2-399. [DOI] [PubMed] [Google Scholar]
- Johnson D. C., Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987 Jul;61(7):2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C., McDermott M. R., Chrisp C., Glorioso J. C. Pathogenicity in mice of herpes simplex virus type 2 mutants unable to express glycoprotein C. J Virol. 1986 Apr;58(1):36–42. doi: 10.1128/jvi.58.1.36-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C., Smiley J. R. Intracellular transport of herpes simplex virus gD occurs more rapidly in uninfected cells than in infected cells. J Virol. 1985 Jun;54(3):682–689. doi: 10.1128/jvi.54.3.682-689.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C., Spear P. G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982 Sep;43(3):1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R., Peitchel R., Goldman J. N., Goldman M. An IgG-Fc receptor induced in cytomegalovirus-infected human fibroblasts. J Immunol. 1976 Mar;116(3):772–777. [PubMed] [Google Scholar]
- Kerbel R. S., Davies A. J. The possible biological significance of Fc receptors on mammalian lymphocytes and tumor cells. Cell. 1974 Oct;3(2):105–112. doi: 10.1016/0092-8674(74)90113-5. [DOI] [PubMed] [Google Scholar]
- Lee G. T., Para M. F., Spear P. G. Location of the structural genes for glycoproteins gD and gE and for other polypeptides in the S component of herpes simplex virus type 1 DNA. J Virol. 1982 Jul;43(1):41–49. doi: 10.1128/jvi.43.1.41-49.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner T., Wilton J. M., Shillitoe E. J. Immunological basis for latency, recurrences and putative oncogenicity of herpes simplex virus. Lancet. 1975 Jul 12;2(7924):60–62. doi: 10.1016/s0140-6736(75)90499-7. [DOI] [PubMed] [Google Scholar]
- Longnecker R., Chatterjee S., Whitley R. J., Roizman B. Identification of a herpes simplex virus 1 glycoprotein gene within a gene cluster dispensable for growth in cell culture. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4303–4307. doi: 10.1073/pnas.84.12.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science. 1987 May 1;236(4801):573–576. doi: 10.1126/science.3033823. [DOI] [PubMed] [Google Scholar]
- MACPHERSON I., STOKER M. Polyoma transformation of hamster cell clones--an investigation of genetic factors affecting cell competence. Virology. 1962 Feb;16:147–151. doi: 10.1016/0042-6822(62)90290-8. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Rixon F. J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985 Jan 5;181(1):1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Moss H. W., McNab D., Frame M. C. DNA sequence and genetic content of the HindIII l region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987 Jan;68(Pt 1):19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- McTaggart S. P., Burns W. H., White D. O., Jackson D. C. Fc receptors induced by herpes simplex virus. I. Biologic and biochemical properties. J Immunol. 1978 Aug;121(2):726–730. [PubMed] [Google Scholar]
- Neidhardt H., Schröder C. H., Kaerner H. C. Herpes simplex virus type 1 glycoprotein E is not indispensable for viral infectivity. J Virol. 1987 Feb;61(2):600–603. doi: 10.1128/jvi.61.2.600-603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikas I., McLauchlan J., Davison A. J., Taylor W. R., Clements J. B. Structural features of ribonucleotide reductase. Proteins. 1986 Dec;1(4):376–384. doi: 10.1002/prot.340010411. [DOI] [PubMed] [Google Scholar]
- Ogata M., Shigeta S. Appearance of immunoglobulin G Fc receptor in cultured human cells infected with varicella-zoster virus. Infect Immun. 1979 Nov;26(2):770–774. doi: 10.1128/iai.26.2.770-774.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. T., Lancet D., Robb R. J., Lopez de Castro J. A., Strominger J. L. The heavy chain of human histocompatibility antigen HLA-B7 contains an immunoglobulin-like region. Nature. 1979 Nov 15;282(5736):266–270. doi: 10.1038/282266a0. [DOI] [PubMed] [Google Scholar]
- Para M. F., Baucke R. B., Spear P. G. Glycoprotein gE of herpes simplex virus type 1: effects of anti-gE on virion infectivity and on virus-induced fc-binding receptors. J Virol. 1982 Jan;41(1):129–136. doi: 10.1128/jvi.41.1.129-136.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para M. F., Goldstein L., Spear P. G. Similarities and differences in the Fc-binding glycoprotein (gE) of herpes simplex virus types 1 and 2 and tentative mapping of the viral gene for this glycoprotein. J Virol. 1982 Jan;41(1):137–144. doi: 10.1128/jvi.41.1.137-144.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson R. H., Bacchetti S., Smiley J. R. Cells that constitutively express the herpes simplex virus immediate-early protein ICP4 allow efficient activation of viral delayed-early genes in trans. J Virol. 1985 May;54(2):414–421. doi: 10.1128/jvi.54.2.414-421.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A. A., Teschner M., Sethi K. K., Brandis H. Appearance of IgG (Fc) receptor(s) on cultured human fibroblasts infected with human cytomegalovirus. J Immunol. 1976 Jul;117(1):253–258. [PubMed] [Google Scholar]
- Rhim J. S., Cho H. Y., Huebner R. J. Non-producer human cells induced by murine sarcoma virus. Int J Cancer. 1975 Jan 15;15(1):23–29. doi: 10.1002/ijc.2910150104. [DOI] [PubMed] [Google Scholar]
- Rosenthal K. L., Smiley J. R., South S., Johnson D. C. Cells expressing herpes simplex virus glycoprotein gC but not gB, gD, or gE are recognized by murine virus-specific cytotoxic T lymphocytes. J Virol. 1987 Aug;61(8):2438–2447. doi: 10.1128/jvi.61.8.2438-2447.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain M. A., Galloway D. A. Herpes simplex virus specifies two subunits of ribonucleotide reductase encoded by 3'-coterminal transcripts. J Virol. 1986 Mar;57(3):802–808. doi: 10.1128/jvi.57.3.802-808.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATKINS J. F. ADSORPTION OF SENSITIZED SHEEP ERYTHROCYTES TO HELA CELLS INFECTED WITH HERPES SIMPLEX VIRUS. Nature. 1964 Jun 27;202:1364–1365. doi: 10.1038/2021364a0. [DOI] [PubMed] [Google Scholar]
- Westmoreland D., St Jeor S., Rapp F. The development by cytomegalovirus-infected cells of binding affinity for normal human immunoglobulin. J Immunol. 1976 Jun;116(6):1566–1570. [PubMed] [Google Scholar]
- Westmoreland D., Watkins J. F. The IgG receptor induced by herpes simplex virus: studies using radioiodinated IgG. J Gen Virol. 1974 Jul;24(1):167–178. doi: 10.1099/0022-1317-24-1-167. [DOI] [PubMed] [Google Scholar]