Fig 9. Crystal Structure of the Protease Domains of FXI and FXIa.
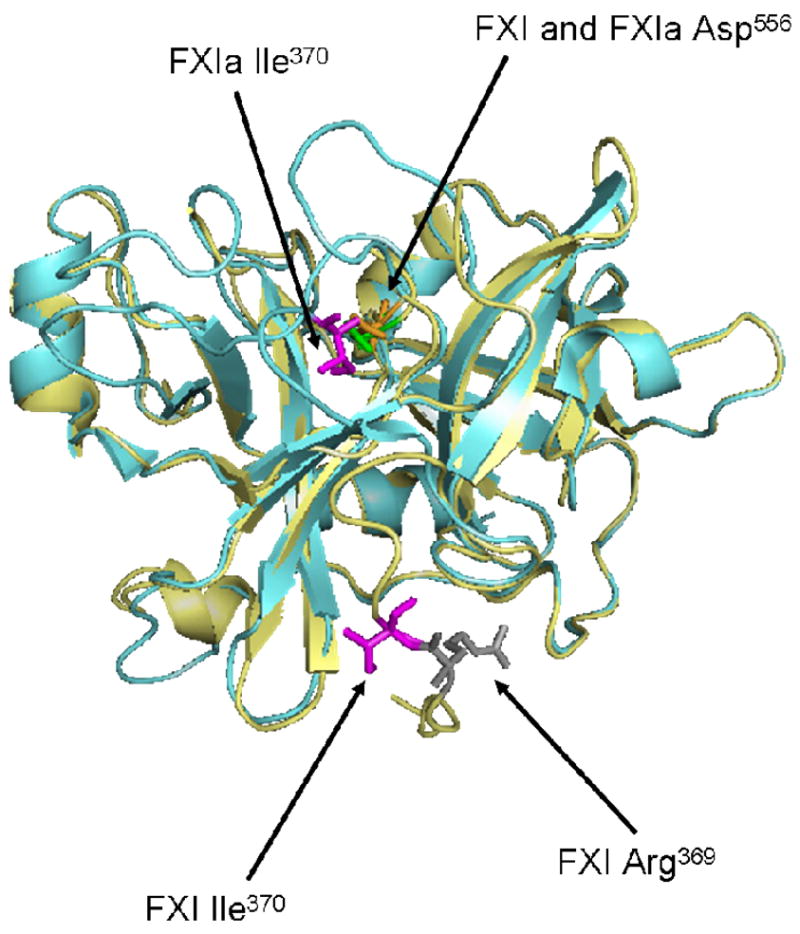
The protease domain of FXIa is shown here superimposed onto the protease domain of the zymogen, FXI. Ile370 (chymotrypsin Ile16) is shown in magenta for both molecules. For FXI, Arg369 is also shown in gray. Upon activation, the scissile bond between Arg369-Ile370 is cleaved and a new N-terminal sequence (Ile-Val-Gly-Gly) is formed. The new N-terminal Ile370 folds inward toward the catalytic triad and forms a salt bridge with Asp194. In the FXIa structure, the major conformational change is the positioning of Ile370, a movement of ~20Å from its placement in the zymogen structure. (PDB:2F83 for zymogen and PDB:1ZJD for enzyme were used to create this model using Pymol software (DeLano Scientific LLC, Palo Alto, CA)).
