Figure 1.
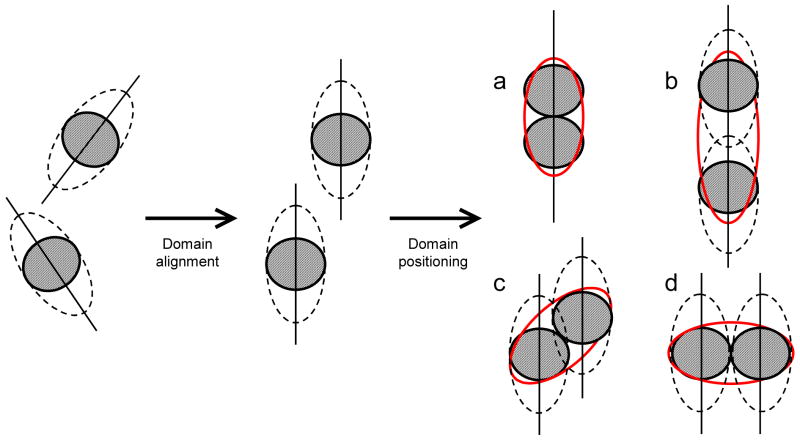
Schematic illustration of the proposed concept of domain positioning based on the rotational diffusion tensor. Provided the structures of the individual domains are available, the interdomain orientation is determined first, and domains are oriented by a rigid-body rotation that aligns the corresponding principal axes (shown as sticks) of the overall diffusion tensor of the complex “reported” by the individual domains, as detailed elsewhere.1,2,10,14 This procedure, however, does not define the relative domain positioning in the molecule. For example, although the interdomain orientation is the same for the domain arrangements (a) through (d), only in (a) is the overall rotational diffusion tensor (red ellipse) consistent with both the magnitude and orientation of the experimental diffusion tensor. In these drawings a dashed ellipse schematically represents the experimentally determined diffusion tensor, whereas a solid red ellipse represents the actual diffusion tensor for a given protein shape/structure. Because the NMR relaxation data sense not only the rate of tumbling but also the orientation of the rotation axes relative to each domain, the relaxation-based approach should be self-sufficient for proper positioning of the domains within a molecule.