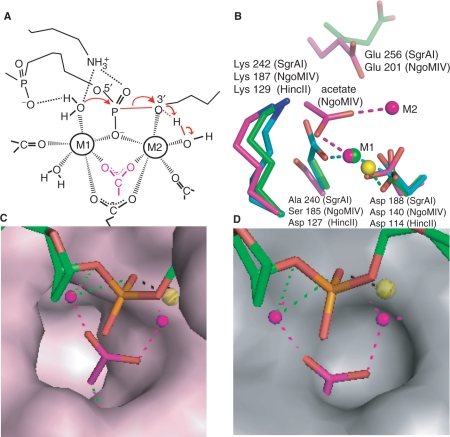
Figure 7.
Comparison of NgoMIV and SgrAI active sites. (A) Model of the two-metal-ion mechanism of DNA cleavage. Red arrows indicate changes in bonding. Scissile P-O3′ bond shown as red line. Group in pink is an acetate molecule in the structure of NgoMIV. (B) Superposition of the active sites of HincII (cyan, blue, red, yellow), SgrAI (green, blue, red) and NgoMIV (magenta, blue, red) using the alpha carbon atoms of the residues shown (with the exception of Glu 256 and Glu 201). The positions of the bound metal ions are shown as spheres at M1 and M2. Interactions between metal ions and ligands are shown as dashed lines. Glu 201 and Glu 256 do not directly ligate metal ions. NgoMIV contains an acetate at roughly the same position as Ser 185, which is Asp in the typical RE motif PDX9–18(E/D)YK. (C) Superposition used in Figure 7B with the solvent accessible surface area of NgoMIV residues shown in pink, and the DNA of the SgrAI Form 2 Mn2+ bound structure shown in green, red and orange. The Mn2+ ion is shown as a yellow sphere, and the Mg2+ ions of the NgoMIV structure shown as pink spheres. The acetate molecule that ligates to both Mg2+ in the NgoMIV structure is shown in pink and red. The acetate fits neatly into a deep pocket in the NgoMIV structure. (D) As in Figure 7C, with the surface of the SgrAI protein shown in grey. The deep pocket found in the NgoMIV surface is absent in SgrAI, and the acetate molecule overlaps with residues of SgrAI forming the surface.