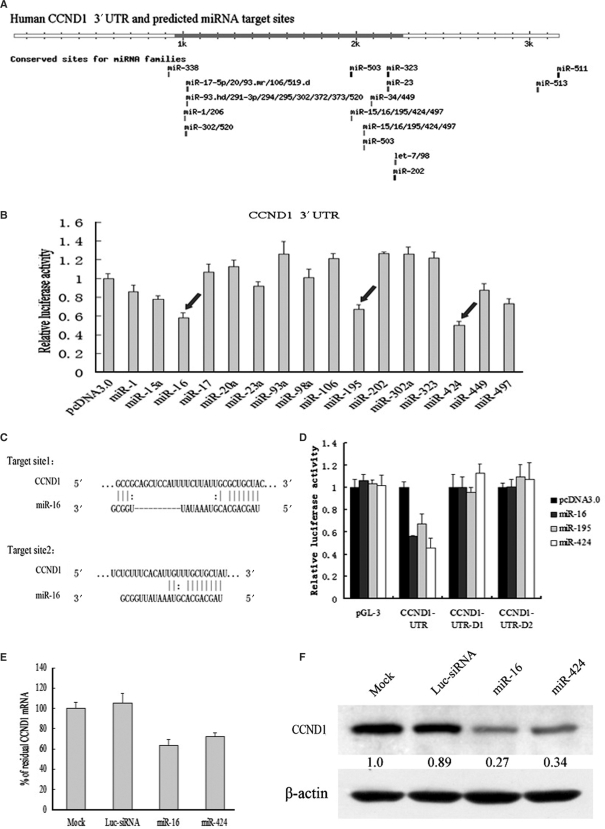
Figure 2.
miR-16 family downregulated CCND1 protein and mRNA by targeting a putative binding site. (A) Human CCND1 3′-UTR and its possible miRNA target sites predicted by targetscan software. Many different miRNA seed regions are complementary to the elements of CCND1 3′-UTR, among which miR-16 family has two conserved sites. (B) Luciferase assays indicated that miR16 family could downregulate the expression of CCND1. pcDNA3.0 control plasmid and 16 different miRNA expressing plasmids were cotransfected with a modified pGL-3 control vector containing CCND1 3′-UTR. Luciferase assays were performed 48 h posttransfection. Histogram shows normalized mean values of relative luciferase activity form three independent experiments. Normalized luciferase activity in the absence of miRNAs expression vector was set to 100%. Arrows indicate a significant reduction of luciferase activity by miR-16/195/424. (C) Sequence inspection by bioinformatics reveals that target site 1(above) and target site 2 (below) are two conserved elements complementary to the seed region of miR-16 (nucleotides 2–8). (D) Luciferase assays indicated that miR-16 family downregulated the expression of CCND1 by targeting putative target site 2. pcDNA3.0 control plasmid, miR-16 expressing plasmid and miR-424 expressing plasmid were cotransfected with a modified pGL-3 control vector containing wild-type CCND1 3′-UTR or two deletants of CCND1 3′-UTR, respectively. The effectiveness of synthetic miR-16/424 on CCND1 mRNA and protein was respectively analyzed by qRT–PCR (E) and western blot (F).