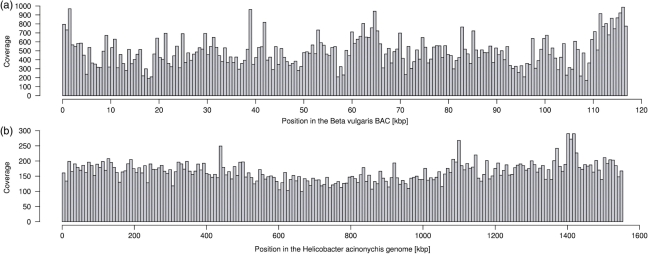
Figure 3.
Distribution of Solexa reads along the reference sequences considering unique match positions reported by ELAND (zero, one or two mismatch bases) and reads with more than one match position (no mismatch bases) detected with a Perl script. (a) Read distribution along the Beta vulgaris BAC sequence (with cloning vector pBeloBACII). 2 166 892 27mer reads were matched against the finished sequence (enclosed by the cloning vector,∼117 kbp in total). The read coverage was calculated in 200 consecutive 0.58 kbp windows. (b) Read distribution along the 1.55 Mbp Helicobacter genome, based on 8 700 113 32mer reads. The local coverage is shown in 200 consecutive windows of 7.77 kbp.