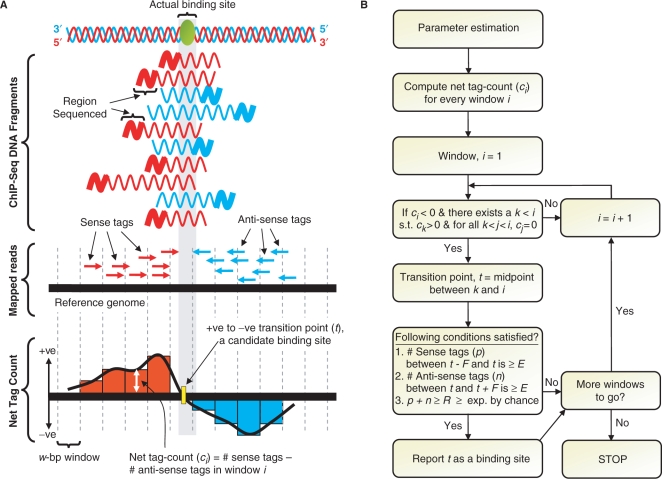
Figure 1.
Schematic overview of SISSRs algorithm. (A) Sequenced short reads (typically ∼25–50 bp) from ChIP-Seq experiments are first mapped onto the reference genome. The mapped reads are then used to estimate statistical parameters, which include the estimation of the average length F of sequenced DNA fragments. (B) The entire reference genome along with mapped reads is scanned using overlapping windows of size w base pairs (overlapping not shown in the figure for clarity), and the net tag count (ci) for every window i is calculated. Every transition point (t) is a candidate binding site, and needs to satisfy a set of estimated as well as user-set thresholds in order to be classified as a true binding site.