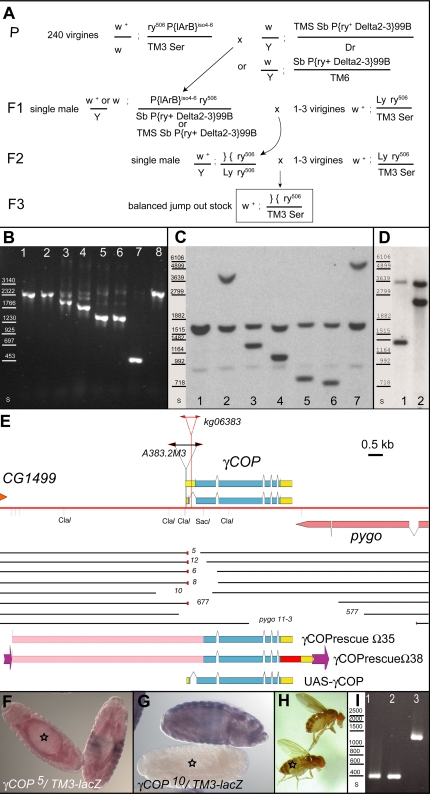
Figure 1. Structure of the γCOP locus and generation of γCOP mutations.
(A) Strategy to generate γCOP jump out excision alleles from γCOPP{lArB}A383.2M3, which carries ry+ as a selection marker. In the parental generation P, the γCOPP{lArB}A383.2M3 line was crossed to any one of the P-element transposase lines, marked with Sb. In the F1 generation, single males undergoing P-element excision events were crossed to virgin Ly ry506 females in order the chromosome, from which the P{lArB} has jumped out (marked }{ ), can be discriminated in the following generations from the homologous chromosome 3. The individual excision events were balanced in the F3 generation (TM3, Ser). (B) In lanes 1–8 PCR amplification products using primers gm4-cop6rev on genomic DNA of control and deletion lines was loaded; primers gm4-cop6rev amplify a fragment of 2305 bp from the γCOP locus of wild type or ry506. (1) ry506, (2) γCOPP{lArB}A383.2M3(iso6)/TM3 Sb (3) γCOP5/TM3 Ser (4) γCOP12/TM3 Ser (5) γCOP6/TM3 Ser (6) γCOP8/TM3 Ser (7) γCOP10/TM3 Ser (8) ry506. Standard molecular weights are indicated in lane s. (C, D) HindIII, EcoRI digested gDNA from control and deletion lines was probed with a DIG-labelled cop5-cop11rev fragment in (C) and with a 3prime1-3prime2rev fragment in (D) (see Materials and Methods); Standard molecular weights of DIG VII are indicated in lane s. The following genotypes were loaded and blotted in (C): ry506 (1), γCOPP{lArB}A383.2M3/TM3 Ser (2), γCOP5/TM3 Ser (3), γCOP12/TM3 Ser (4), γCOP6/TM3 Ser (5), γCOP8/TM3 Ser (6), γCOP10/TM3 Ser (7). In (D) γCOP577/TM3 Ser (1), γCOP677/TM3 Ser (2). (E) Map of γCOP locus and the γCOPP{lArB}A383.2M3 deletions. Chromosome 3R is indicated as a red line. P-element insertion of γCOPP{lArB}A383.2M3 is indicated in black, γCOPkg06383 insertion in red. Extent of the two alternative γCOP transcripts is shown, yellow boxes denote non-coding regions, blue boxes coding regions. cDNA LP01448 was used for cloning experiments (Materials and Methods). The 3'end of the neighboring gene CG1499 is indicated above the red line. The 3'end of the neighboring gene pygo is indicated below the red line. The DNA present on the deletion chromosomes is indicated as black lines; the missing DNA in comparison to the original chromosome is a blank space; small red triangles indicate the parts of the P-element inverted repeat which stayed behind after P-element excision. In deletion γCOP10 and Df(3R)γCOP577 no P-element derived sequences have stayed behind. Constructs for transgenic flies are shown. The extent of the upstream genomic region present in rescue construct Ω35 and Ω38 is shown as a pink box; Ω38 is tagged with mRFP (red box) and the γCOP 3'UTR (yellow box); in addition this construct is flanked by FRT sites (purple arrows); in UASp-γCOP the full length γCOP cDNA is present (see Materials and Methods). (F, G) Deletion alleles (balanced over TM3-lacZ) were analyzed for γCOP transcription with a DIG-labeled γCOP probe and a FITC-labeled βGal probe, to discriminate heterozygous (red and blue) from homozygous γCOP mutant embryos. Whereas deletion mutant γCOP5 (marked with asterisk in (F)) still expresses γCOP (red staining), deletion mutant γCOP10 (marked with asterisk in (G)) shows no γCOP transcripts. (H) Fly rescued with γCOP rescue construct (marked with asterisk) and heterozygous sibling fly (unmarked) are shown. (I) PCR amplification of primer gm4 and cop6rev on five rescued flies confirming the presence of the original deletion in the rescued viable adults; in the PCR reaction only the short amplicon of the deletion breakpoint is visible; Lane (s) shows standard sizes. The amplicon of γCOP mutant flies (γCOP10 (1,2) γCOP6 (3)), rescued with insertion Ω35-i7 (2) or insertion Ω35-i8 (2, 3).