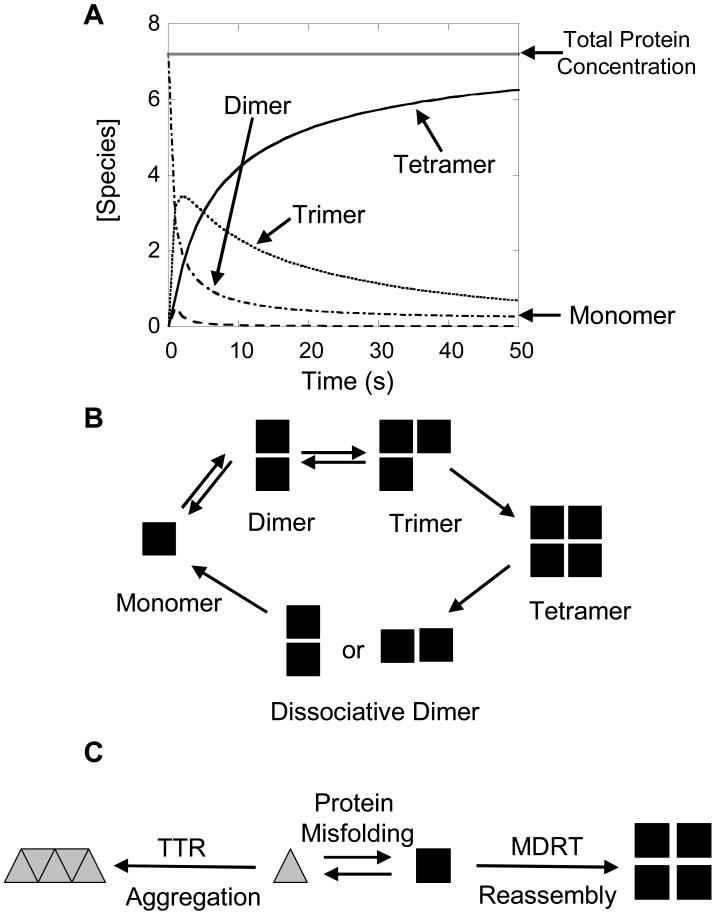
Figure 8.
Graphic representation of the kinetic partitioning of TTR monomers. A) Kinetic plot of the concentration of monomer (- - - -), dimer (- • - • -), trimer (••••), and tetramer (solid line) during the reassembly of TTR tetramer ([Protein]tot = 7.2 µM) as predicted by the rate constants determined by the fitting analysis (Table 1C). The dotted gray line indicates the total protein concentration representing the predicted amplitude upon completion of the reassembly reaction (7.2 µM). The initial rate of TTR tetramer formation is fast because of the high concentration of monomer in solution. As the reaction proceeds to completion, the rate of tetramer formation becomes slower because of the depleted concentration of TTR monomer in solution. B) TTR tetramers are formed by the sequential addition of monomers to dimeric and trimeric intermediates along the reassembly pathway. The tetramer dissociates in a cooperative unfolding pathway through a dimeric intermediate. The dimeric intermediate along the dissociative pathway may be the same unstable dimer formed in the reassembly mechanism, explaining the cooperative unfolding of this species under denaturing conditions. C) TTR monomers can kinetically partition between the reassembly pathway (black symbols) and the protein aggregation pathway (gray symbols). The two mechanistic pathways are buffered by a protein misfolding step. The partitioning of TTR into these two pathways can dictate the rate and extent of TTR aggregation, potentially affecting disease pathology.