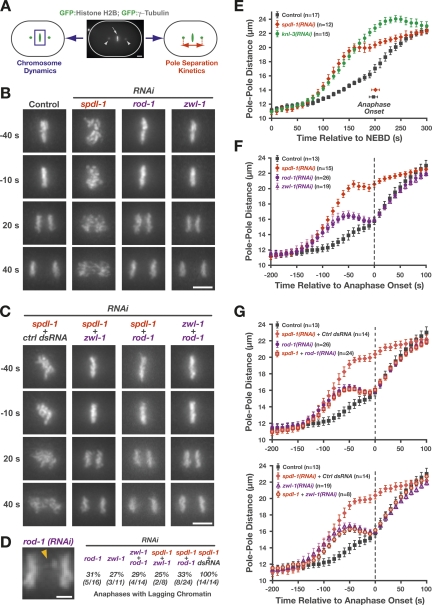
Figure 6.
Depletion of SPDL-1 results in a more severe chromosome segregation defect than depletion of ROD-1 or ZwilchZWL-1. (A) Cartoon outlining the two parameters monitored in a strain expressing GFP:histone H2B and GFP:γ-tubulin: chromosome dynamics and kinetics of spindle pole separation. (B) Frames from time-lapse sequences of the first embryonic division, highlighting the differences in chromosome dynamics after depletion of SPDL-1 and the RZZ complex subunits ROD-1 and ZwilchZWL-1 (see also Supplemental Movie 11). The time point 0 sec denotes the onset of sister chromatid separation. Bar, 5 μm. (C) Selected frames from time-lapse sequences of embryos codepleted of RZZ subunits and SPDL-1, which significantly reduces the severe chromosome missegregation phenotype of SPDL-1 single depletions to match that of RZZ subunit single depletions (see also Supplemental Movie 12). Bar, 5 μm. (D) Representative image of anaphase with lagging chromatin in a rod-1(RNAi) one-cell embryo. The frequency of one-cell embryos with lagging anaphase chromatin is indicated for single and double inhibitions involving RZZ subunits and SPDL-1. Bar, 2 μm. (E) Pole separation kinetics in wild-type, spdl-1(RNAi), and knl-3(RNAi) embryos. Images were acquired at 10-sec intervals, and sequences were time-aligned relative to NEBD. Pole–pole distances in the time-aligned sequences were measured, averaged for the indicated number (n) of embryos, and plotted against time. Error bars represent the SEM with a 95% confidence interval. (F) Pole separation kinetics of the perturbations shown in B. Sequences were time-aligned relative to the onset of sister chromatid separation (“Anaphase Onset”). Error bars represent the SEM with a 95% confidence interval. (G) Pole separation kinetics of the perturbations shown in C, demonstrating that double depletions of SPDL-1 and RZZ complex subunits result in a pole separation profile that is indistinguishable from RZZ subunit single depletions. For controls, spdl-1 dsRNA was diluted equally with dsRNA corresponding to the budding yeast gene CTF13 or the C. elegans gene sas-5, which is not required for the first embryonic division (both conditions gave identical results). Error bars represent the SEM with a 95% confidence interval.