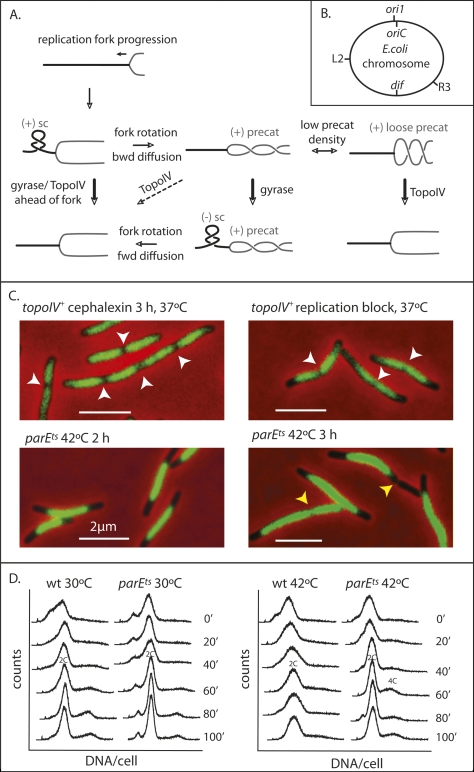
Figure 1.
TopoIV impairment prevents sister nucleoid separation. (A) Schematic of topological processing during DNA replication. As a replication fork moves forward, the DNA ahead of the fork becomes overwound and the density of (+) RH linkage increases, forming (+) supercoils (sc) with a LH plectonemic superhelix ahead of the fork. The torsional stress so generated is relieved if rotation of the fork replisome occurs, thereby redistributing the RH linkage. This leads to formation of (+) RH precatenanes behind the fork. These are only good substrates for TopoIV if their density is low (“loose” precat), so their crossing angle can become optimal for TopoIV action (Stone et al. 2003). If precatenane-containing DNA becomes underwound ahead of the fork, generating (−) sc, then diffusion forward of RH precatenanes to remove (−) sc will be favored. It has also been proposed that accumulation of (−) sc ahead of a fork can lead to LH (−) precatenanes in newly replicated DNA, with these also being an in vivo substrate for TopoIV (see Schvartzman and Stasiak 2004). We believe this is unlikely to be a common occurrence, because it would exacerbate chromosome unlinking and be energetically unfavorable because ATP hydrolysis by gyrase ahead of a fork, generating (−) supercoiling, would need to be countered by ATP hydrolysis by TopoIV to remove LH precatenanes. Unreplicated DNA is shown as a thick black line and newly replicated DNA is shown as a gray line. Arrows indicate the direction of reaction. Thick arrows indicate efficient reactions and dashed arrows inefficient reactions. (B) Schematic of the E. coli genetic loci used. (C) Nucleoids in the indicated cells. Replication was blocked by TetR binding to tetO arrays at R3, ∼700 kb anti-clockwise of dif. White arrows indicate positions of nucleoid splitting and yellow arrows show positions of cytokinesis in a bright-field image. Nucleoids were stained with DAPI (0.5 μg/mL). Bars, 2 μm. (D) Flow cytometry (DNA content per cell) of wild-type and parEts cells grown at 30°C and 42°C. Rifampicin, which prevents replication initiation, and cephalexin, which blocks cytokinesis, were added at 0 min, and samples were taken at the indicated times.