Summary
This study was undertaken to further elucidate the biological mechanisms underlying the frequent event of transformation of follicular lymphoma (FL) to diffuse large B-cell lymphoma (t-FL). The gene expression profiles of 20 paired lymph node biopsies, derived from the same patient pre- and post-transformation, were analysed using the Lymphochip cDNA microarray. TP53 mutation analysis was performed and copy number alterations at the c-REL and CDNK2A examined. Immunohistochemistry was performed on an independent panel of paired transformation paraffin-embedded samples. Transformed follicular lymphoma was predominantly of the germinal centre B-like phenotype both at the mRNA and protein level. Despite this homogeneity, transformation proceeded by at least two pathways. One mechanism was characterised by high proliferation, as assessed by the co-ordinately expressed genes of the proliferation signature. This group was associated with the presence of recurrent oncogenic abnormalities. In the remaining cases, proliferation was not increased and transformation proceeded by alternative routes as yet undetermined. Genes involved in cellular proliferation prevailed amongst those that were significantly increased upon transformation and T cell and follicular dendritic-associated genes predominated amongst those that decreased. t-FL is a germinal centre B (GCB)-like malignancy that evolves by two pathways, one that is similar in proliferation rate to the antecedent FL and the other that has a higher proliferation rate and is characterised by the presence of recognised oncogenic abnormalities.
Keywords: follicular lymphoma, transformation, diffuse large B-cell lymphoma, expression profiling
Transformation of follicular lymphoma (FL) to diffuse large B-cell lymphoma (DLBCL, t-FL) occurs commonly (Cullen et al, 1979; Hubbard et al, 1982; Garvin et al, 1983; Horning & Rosenberg, 1984; Bastion et al, 1997), being generally associated with a poor prognosis (Armitage et al, 1981) despite the use of adjunctive high-dose therapy in selected individuals (Bastion et al, 1997; Chen et al, 2001; Williams et al, 2001). Further biological insights and new therapeutic strategies are therefore urgently required if the outlook for these patients following transformation is to be improved.
The comparison of biopsy material from the same individual pre- and post-transformation is a prerequisite for understanding the molecular mechanisms driving the phenotypic change of lymphoma. To date, studies of this kind have revealed a number of recurring cytogenetic abnormalities (Goff et al, 2000; Hough et al, 2001; Boonstra et al, 2003; Martinez-Climent et al, 2003); mutation of TP53 (Lo Coco et al, 1993; Sander et al, 1993; Davies et al, 2005), inactivation of CDKN2A and CDKN2B (Elenitoba-Johnson et al, 1998) and c-MYC rearrangement (Yano et al, 1992) or mutation (Lossos et al, 2002). These secondary events appear to drive transformation in small subsets of patients. Other acquired insults have also been recognised, including amplification (Nagy et al, 2000; Martinez-Climent et al, 2003) and mutation (Matolcsy et al, 1997) of the translocated BCL2, translocations (Akasaka et al, 2003) or mutation of the 5′ UTR of BCL6 (Capello et al, 2000; Lossos & Levy, 2000; Szereday et al, 2000) and c-REL amplifications (Goff et al, 2000). Each individual lesion occurs with a relatively low frequency, and serves to exemplify the diversity of the biological process. The limited number of gene expression studies performed on cryopreserved sequential biopsy tissue (Lossos et al, 2002; Elenitoba-Johnson et al, 2003; de Vos et al, 2003; Glas et al, 2005), has added to our understanding of the transformation event, yet a lack of concordance between results is observed, a probable reflection of the small number of paired samples available in each study population.
In the light of such heterogeneity, we collated the largest series of paired samples for array-based gene expression profiling, supplemented with genomic analysis and immunohistochemical (IHC) evaluation to gain additional new insights into the mechanisms of FL transformation.
Patients and methods
Patient selection
Fresh snap frozen lymph node biopsy material was retrieved from the tissue archive of the four contributing institutions (St Bartholomew’s Hospital, University of Nebraska Medical Centre, The Norwegian Radium Hospital and Southwest Oncology Group). Each Institution’s respective Institutional Review Board or Local Ethics Committee approved the study. Inclusion required the availability of both a sample of FL (grade 1, 2 or 3a) and a subsequent sample of DLBCL obtained at the time of transformation from the same patient. Paraffin-embedded sections from each biopsy were reviewed by the Pathology Committee of the Leukemia and Lymphoma Molecular Profiling Project. Twenty pairs were available for final analysis: seven grade 1 FL to DLBCL, nine grade 2 FL to DLBCL and three grade 3a FL to DLBCL. Consensus was not reached in the pathological grading of one FL sample. Analysis was also carried out in five cases of t-FL where no antecedent FL sample was available for gene expression profiling, however previous FL had been confirmed by review of paraffin-embedded material.
Gene expression profiling
Lymphochip cDNA microarrays were used to quantify mRNA expression according to previously described methodology (Alizadeh et al, 2000; Rosenwald et al, 2002). A list of genes that significantly differed between FL and DLBCL was constructed using the random variance model t-test (Wright & Simon, 2003), with the cut-off value for P set at <0.001. The probability of belonging to a particular sub-class of DLBCL was determined using the Bayesian statistical predictor and clone set previously reported (Wright et al, 2003).
Genomic gains/losses and TP53 mutation status
Genomic copy number changes were investigated using real-time polymerase chain reaction (PCR) assays for two established oncogenes/tumour suppressors reported to be of putative significance in transformation [c-REL (Goff et al, 2000) and CDKN2A (Labuhn et al, 2001; Rosenwald et al, 2003)]; in each case β2 microglobulin was used as the control gene (Goff et al, 2000). In addition, the entire coding sequence of TP53 was screened for the presence of mutations by PCR/single stranded conformation polymorphism analyses (Davies et al, 2005). Loss of heterozygosity (LOH) at the TP53 locus was investigated by studying the pattern of five common polymorphisms. Paired sample clonality was determined by a PCR-based IGH gene rearrangement assay with fluorescence detection (InVivoScribe Technologies, San Diego, CA, USA) and IGH/BCL2 status by PCR across the major breakpoint region (Iqbal et al, 2004a) and minor cluster region (Price et al, 1991; Leonard et al, 1998) breakpoints (Table SI).
Tissue microarray (TMA) construction and immunohistochemistry
An independent validation set of 35 patients with at least one pre- and post-transformation paraffin-embedded biopsy sample available was derived from the Pathology archive of St Bartholomew’s Hospital. Full-sized 5 μm haematoxylin and eosin sections were marked and 1-mm diameter cores taken in triplicate from the paraffin-embedded tissue using a manual arrayer (Beecher Instruments, Sun Prairie, WI, USA). All cores from a single patient were randomly included on the same TMA with six control cores of reactive lymphoid tissue. Four TMAs accommodated all samples (collectively the Tx-TMA). To ensure Tx-TMA cores accurately represented the corresponding full tissue sections (n = 10) a panel of nine routine IHC diagnostic markers was scored. Concordance was >90%. A second TMA, comprising 31 de novo DLBCL samples, was produced for methodological validation.
Immunohistochemical analysis was carried out as previously described (Dahia et al, 1998) using the following antibody panel: MUM1/IRF4, BCL-6 (DakoCytomation, Ely, UK) and CD10 (Novocastra Laboratories Ltd, Newcastle Upon Tyne, UK). To distinguish germinal centre B (GCB)-like DLBCL from non-GCB like DLBCL, the MUM1/IRF4, BCL-6 and CD10 panel was scored as previously reported by Hans et al (2004). The entire TMA cores were analysed by two independent pathologists, and the number of positively stained cells was expressed as an average percentage.
Results
The common clonal origin of 18/20 pairs of FL and t-FL was confirmed by PCR based IGH and IGH/BCL2 rearrangement studies (Table SI). Initially, the molecular subtype of t-FL was classified according to the GCB-like or activated B-cell (ABC)- like cell of origin model of de novo DLBCL. Using a Bayesian predictor (Wright et al, 2003), all classifiable cases (18/25) of t-FL cases were of GCB-like phenotype; the remaining seven samples were ‘unclassifiable’ by the predictor (Fig 1). No t-FL samples were of the ABC-like phenotype. Using an independent validation set of 35 FL and t-FL patient pairs, a Tx-TMA was generated from paraffin-embedded tissue. Using the pattern of CD10, BCL6 and MUM1/IRF4 expression that was previously reported to discriminate between GCB and non- GCB de novo DLBCL (Hans et al, 2004), 89% (31/35) of t-FL was of the GCB phenotype (28/35 (80%) CD10+ and 3/35 (9%) CD10−, BCL6+, MUM1−). The remaining, 3/35 (9%) were classified as non-GCB phenotype (CD10−, BCL6+, MUM1+) with one unclassifiable. In a parallel control TMA generated to validate the methodology, and comprising 31 cases of de novo DLBCL, 35% of samples were of GCB phenotype (5-year survival, 73%) and 65% were non-GCB (5- year survival, 51%).
Fig 1.
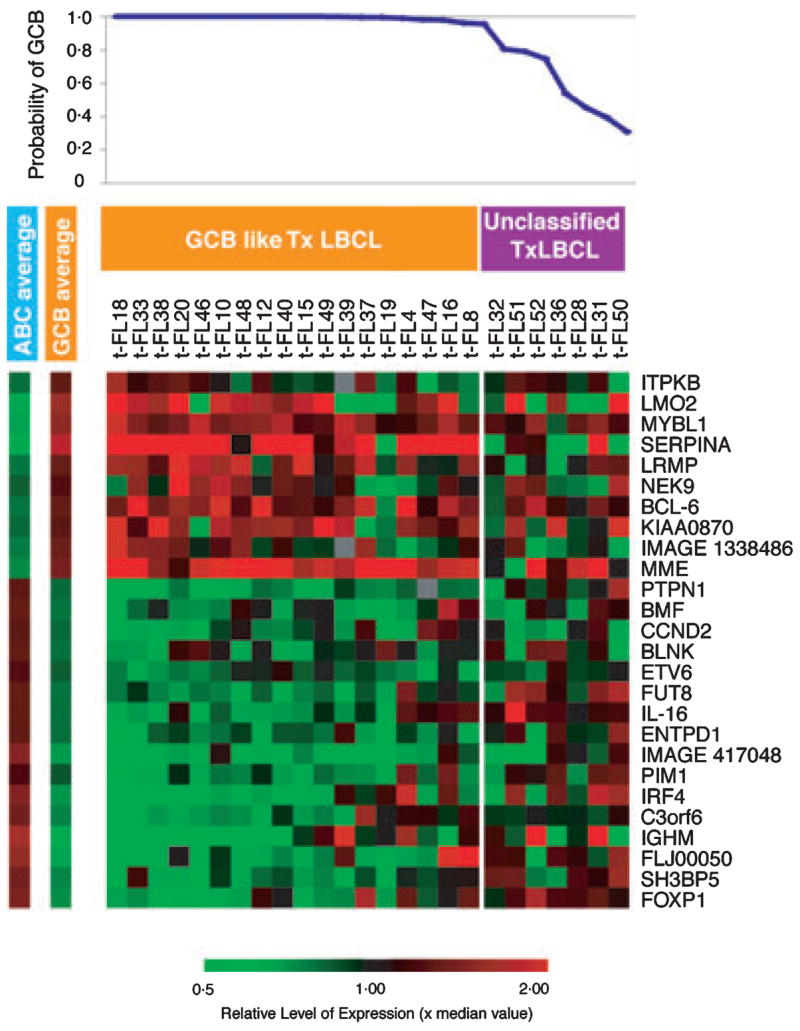
Sub-group predictor analysis to classify transformed follicular lymphoma (FL). The expression of 26 genes previously identified in de novo diffused large B-cell lymphoma (DLBCL) to classify tumour according to the germinal centre B-cell or activated B-cell-like subgroup was used to analyse the phenotype of DLBCL arsing from FL (t-FL). The average expression of genes comprising the predictor in each sub-group are shown (left). The probability of any sample belonging to a sub-group was determined and, if >0.9, was considered as member of that class.
Seventy-six genes demonstrated a highly significant (P < 0.001) difference in expression between FL and t-FL using a supervised approach (Fig 2). No single gene demonstrated a uniform pattern of expression change across all samples upon transformation. Genes involved in cellular proliferation predominated amongst the 29 that were up-regulated; in addition, genes involved in metabolism, cytoskeletal components, molecular chaperones and apoptosis were also identified. Of the 47 genes with significantly reduced expressions upon transformation, T-cell-associated or follicular dendritic cell genes were in the majority.
Fig 2.
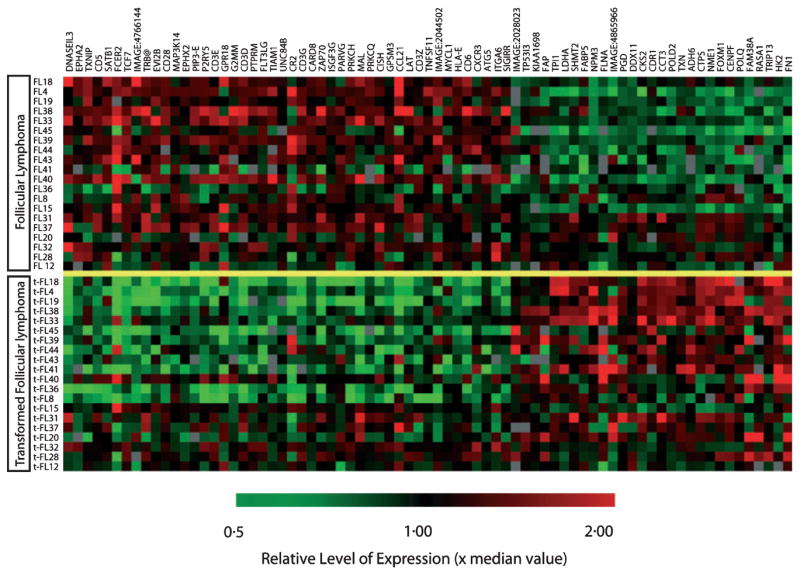
Differential gene expression in supervised analysis between paired follicular lymphoma and transformed follicular lymphoma (t-FL) samples. From 118 clones, 76 genes represented by distinct UniGene clusters were highly significant with a P-value of <0.001 between different histological phenotypes. The gene identification is annotated to the right. Genes involved in cellular proliferation (POLQ, POLD2, CENPF, CTPS, NME1, FOXM11, DDX11, SHMT2, TXN, CCT3, TPI1 and NPM3) predominated amongst those up regulated upon transformation. In addition, genes involved in metabolism (HK2, PGD, FABP5 and LDHA), cytoskeletal components (FN1 and FLNB), molecular chaperones (CCT and NPM3) and apoptotic genes (TXN, TP53I3 and HK2) were also up regulated in t-FL. T cell associated (CD3E, CD3G, CD6, CD28, CXCR3, LAT, TRB@, PRKCQ, MAL, ZAP70, FLT3LG, TCF7, G2MM and DNASEIL3) and follicular dendritic cell genes (CR2 and FCER2) predominated amongst genes that demonstrated significantly reduced expressions upon transformation.
Fourteen of the 29 (48%) genes that demonstrated increased expression upon transformation belonged to a set of coordinately regulated genes expressed during cell division, known as the proliferation signature (Shaffer et al, 2001; Rosenwald et al, 2003). The difference in all proliferation signature genes between t-FL and the antecedent FL sample was examined, and in each case an expression signature average generated. Surprisingly, the proliferation signature was not globally up-regulated in all samples upon transformation (Fig 3). Indeed, only 10/20 pairs showed a significant increase in expression of these genes (mean change >0.5 log expression). In the remainder, the change was minimal or actually decreased, confirming that transformation must proceed by more than one distinct pathway.
Fig 3.
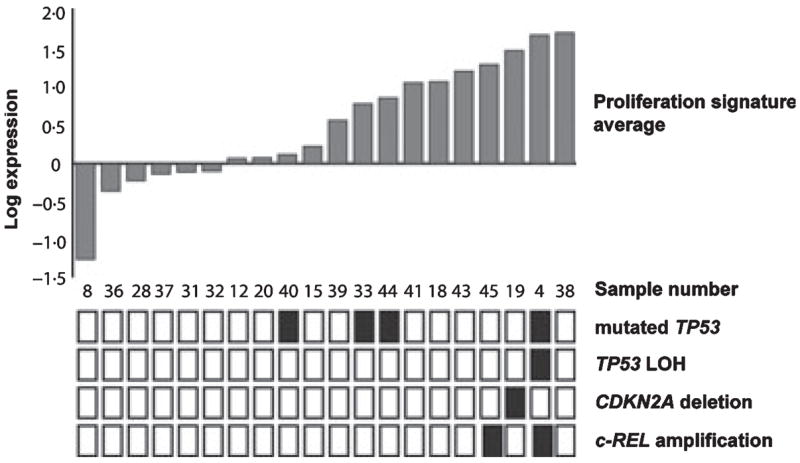
Average change in proliferation signature genes upon transformation and occurrence of genomic aberrations in each transformed follicular lymphoma (t-FL) sample. For 20 genes, comprising the proliferation signature, the change in expression between follicular lymphoma and t-FL was calculated for each sample pair. The average log change was then determined for all clones in each pair. Black squares indicate the presence of either TP53 mutation, loss of heterozygosity and the TP53 locus, CDKN2A loss or c-REL amplification. Clear boxes represent wild type configuration.
To further substantiate this observation, single gene genomic lesions associated with transformation were examined in this series. Mutations in TP53 were observed in 20% (4/20) t-FL samples, with wtTP53 in the antecedent FL sample documented in all but one pair (Table I). LOH at the TP53 locus was observed in only one t-FL sample. The c-REL oncogene was amplified in two patients (in one case the amplification was present prior to histological transformation), while CDKN2A deletion was present in only a single t-FL sample. In all but one case these insults were mutually exclusive (Fig 3). These genomic lesions were observed primarily in those samples demonstrating an increase in expression of proliferation signature genes upon transformation.
Table I.
TP53 mutation acquisition upon transformation of follicular lymphoma (FL) to transformed follicular lymphoma (t-FL).
| Patient | Histology | Nucleotide change | Codon | Exon | Amino acid change | LOH | Functional significance |
|---|---|---|---|---|---|---|---|
| 4 | FL | wt | |||||
| t-FL | 13344 T>C | 584 | 6 | I195T | + | NA | |
| 33 | FL | wt | |||||
| t-FL | 13338 A>T | 578 | 6 | H193L | NA | ||
| 40 | FL | wt | |||||
| t-FL | 14060 G>A | 733 | 7 | G245S | Dominant negative (Shi et al, 2002) | ||
| 44 | FL | 13203 G>A | 521 | 5 | R175H | Dominant negative (Kern et al, 1992; Marutani et al, 1999) | |
| t-FL | 13203 G>A | 521 | 5 | R175H |
Nucleotide numbers refer to the reference sequence (GenBank accession number U94788). Loss of heterozygosity (LOH) was examined at five common polymorphic sites: 11827 G>C, 11992 C>A, 11951_11952ins16, 12139 G>C and 17708 A. Two of these mutations have previously been reported (Davies et al, 2005).
NA, no functional data available; wt, wild type.
The expression of c-MYC (measured by three distinct clones with high concordance) and its target genes were examined, given that it has previously been reported to segregate t-FL into distinct classes (Lossos et al, 2002). In this series a marked increase in c-MYC expression between FL and t-FL was observed in 70% (14/20) pairs, although it did not feature amongst the most significant gene list. Changes in c-MYC expression upon transformation correlated closely with that of 11 direct c-MYC target genes (r = 0.8188, P = 2.65 × 10−5) (selected as direct human c-MYC targets based upon strong supporting evidence) (Table SII). However, the expression of c-MYC and the target genes correlated closely with that of the entire proliferation signature average (r = 0.8398, P = 1.26 × 10−5 and r = 0.975, P = 1.77 × 10−8 respectively) (Fig 4). Thus, dysregulation of c-MYC and its target genes during transformation coincides with a general increase in proliferation rate, as measured by the proliferation signatures and is not the single responsible force driving uncontrolled proliferation nor necessarily the initiating oncogenic event in this subgroup of patients.
Fig 4.
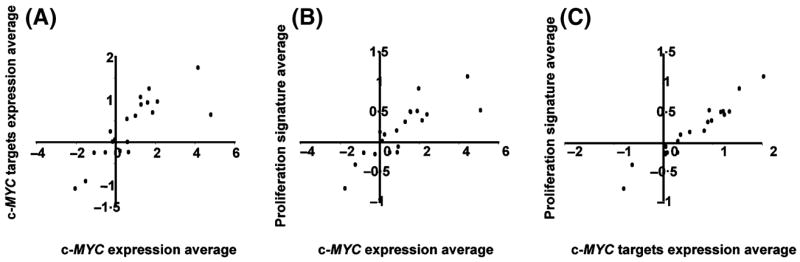
Correlation of expression of c-MYC, c-MYC target genes and the proliferation signature average. (A) Tight correlation between c-MYC expression signature average and c-MYC target genes was observed, similarly for (B) c-MYC and proliferation signature average and (C) proliferation signature to c-MYC target genes.
Discussion
This study, the largest of its kind, sheds new light on the biological and molecular features of FL transformation. First, t-FL has a gene expression phenotype similar to GCB DLBCL and never adopts the ABC DLBCL phenotype. Secondly, transformation of FL occurs by two predominant pathways, one characterised by higher proliferation and the other not. These two pathways differ in all aspects of cellular proliferation, not just in the expression of c-MYC and its target genes. Thirdly, we discovered that recurrent oncogenic events in FL transformation – namely TP53 mutation, CDNK2A loss and c-REL amplification – occur exclusively during transformation by the ‘proliferation’ pathway.
There is remarkable homogeneity in the biological phenotype of t-FL. Such predominance of the GCB-like phenotype sits well with the distinct biological ‘cell of origin’ theory, which defines molecular subclasses of DLBCL indicative of different functional stages of B-cell development (Alizadeh et al, 2000; Rosenwald et al, 2002). IGH/BCL2 translocations are observed almost exclusively in the GCB-like group of de novo DLBCL (Rosenwald et al, 2002; Iqbal et al, 2004b). It is perhaps intuitive, based upon the germinal centre origin of FL, that t-FL should be GCB-like. Such modelling in de novo DLBCL also carries prognostic significance (Alizadeh et al, 2000; Rosenwald et al, 2002). Although the GCB-like class exhibits a more favourable outcome, histological transformation is diagnosed at the time of disease recurrence where the prognostic value of GCB/ABC identity is unknown.
Simultaneous analysis of gene expression and genomic lesions demonstrate that aberration of at least two distinct molecular pathways result in phenotypic transformation. An increase in tumour proliferation rate, as reflected by the expression of proliferation signature genes, occurred in a subgroup of patients. This subgroup of cases was enriched, in a mutually exclusive fashion, for the presence of previously recognised oncogenic events. TP53 loss and mutation, CDNK2A loss and c-REL amplification all appeared to exert a similar end effect, to increase the proliferate rate of the tumours. The mechanism underlying transformation in the non-proliferate sub-groups is intriguing and, despite an extensive search of expression changes, we were unable to present a unifying hypothesis to underpin this subgroup.
Given that expression of c-MYC and its target genes had been previously identified as a classifier and of potential mechanistic importance in t-FL (Lossos et al, 2002), we explored further its contribution to transformation in this dataset. Whilst changes in c-MYC expression were variable upon transformation, and mirrored in a rigorously defined set of c-MYC target genes, our study did not support dysregulation of c-MYC as a primary mechanism. c-MYC and expression of its targets simply followed the pattern of the proliferation signature as a whole and in this conception, thus serves only as a surrogate for the entire proliferation signature, and the multiple oncogenic embraced by it. This is further emphasised by the low frequency of translocations (Yano et al, 1992; Lo Coco et al, 1993), amplifications (Martinez-Climent et al, 2003) and mutations (Lossos et al, 2002) in c-MYC documented in association with transformation.
Beyond our observations, this study clarifies the findings of other investigators who have recognised increased expression of LDHA, often employed as a biochemical surrogate marker, as highly significant upon transformation (Lossos et al, 2002; Glas et al, 2005). With a sample population two-thirds larger than any previous investigation, we report a significant gene list with a 15% identity to the 76 genes employed successfully to distinguish clinically ‘aggressive’ from ‘indolent’ FL (Glas et al, 2005) (of 69 genes present on both platforms, HK2, CENPF, CTPS, LDHA, P2RY5 and DNASE1L3 were common to both series). The role of these genes warrant further exploration, yet such similarity is contrasted with only minimal identity to other datasets (Elenitoba-Johnson et al, 2003; de Vos et al, 2003). Enhanced expression of the glycolytic genes (HK2, PGM and LDHA) reflects a shift in energy production from oxidative phosphorylation to aerobic glycolysis (the Warburg effect) so providing a clear growth advantage (Gatenby & Gillies, 2004) and is the basis of the increased 18F-fluorodeoxyglucose avidity observed by positron emission tomography post-transformation (Schoder et al, 2005). In addition, HK2 binds Bax and inhibits cytochrome c release from the mitochondria, impeding apoptosis (Pastorino et al, 2002). Pharmacological inhibition of HK2 appears feasible and presents a therapeutic target which is already being explored in other malignancies (Geschwind et al, 2004).
In conclusion, significant gene expression changes have been identified upon transformation of FL to t-FL, which, despite their heterogeneous nature, help to further elucidate the biological mechanisms underlying this event. Much remains still to be uncovered, particularly in relation to the subgroup that transforms independently of an increase in proliferation signature genes, but defining distinct subsets of patients with transformed FL and correlating to clinical behaviour may provide a better model for future selection of appropriate prediction, prognostic and targeting strategies.
Acknowledgments
We are grateful to Michael Chiorazzi, Hong Zhao, Sarah Henrickson and Andrew Clear for their expert technical assistance. Also to Dr C Taylor, Mutation Detection Unit, St James’s Hospital, Leeds for TP53 screening and Dr E Birt, Molecular Haematology, Barts and The London School of Medicine and Dentistry for assistance with IGH rearrangement real-time quantitative PCR. The research was supported in part by the Intramural Research Programme of the National Institutes of Health, National Institute, Center for Cancer Research, USA and Cancer Research UK. A.J.D was in receipt of the McElwain Scholarship from the Association of Cancer Physicians. A.R. is supported be the Interdisciplinary Center for Clinical Research (IZKF), University of Würzburg.
Footnotes
Supplementary material
The following supplementary material is available for this article online:
Appendix SI. References.
Table SI. Common clonal origin of paired samples determined by PCR of IGH and IGH/BCL2 rearrangements.
Table SII. Direct c-MYC target genes.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Akasaka T, Lossos IS, Levy R. BCL6 gene translocation in follicular lymphoma: a harbinger of eventual transformation to diffuse aggressive lymphoma. Blood. 2003;102:1443–1448. doi: 10.1182/blood-2002-08-2482. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Armitage JO, Dick FR, Corder MP. Diffuse histiocytic lymphoma after histologic conversion: a poor prognostic variant. Cancer Treatment Reports. 1981;65:413–418. [PubMed] [Google Scholar]
- Bastion Y, Sebban C, Berger F, Felman P, Salles G, Dumontet C, Bryon PA, Coiffier B. Incidence, predictive factors, and outcome of lymphoma transformation in follicular lymphoma patients. Journal of Clinical Oncology. 1997;15:1587–1594. doi: 10.1200/JCO.1997.15.4.1587. [DOI] [PubMed] [Google Scholar]
- Boonstra R, Bosga-Bouwer A, Mastik M, Haralambieva E, Conradie J, van den Berg E, van den Berg A, Poppema S. Identification of chromosomal copy number changes associated with transformation of follicular lymphoma to diffuse large B-cell lymphoma. Human Pathology. 2003;34:915–923. doi: 10.1016/s0046-8177(03)00350-2. [DOI] [PubMed] [Google Scholar]
- Capello D, Vitolo U, Pasqualucci L, Quattrone S, Migliaretti G, Fassone L, Ariatti C, Vivenza D, Gloghini A, Pastore C, Lanza C, Nomdedeu J, Botto B, Freilone R, Buonaiuto D, Zagonel V, Gallo E, Palestro G, Saglio G, Dalla-Favera R, Carbone A, Gaidano G. Distribution and pattern of BCL-6 mutations throughout the spectrum of B-cell neoplasia. Blood. 2000;95:651–659. [PubMed] [Google Scholar]
- Chen CI, Crump M, Tsang R, Stewart AK, Keating A. Autotransplants for histologically transformed follicular non- Hodgkin’s lymphoma. British Journal of Haematology. 2001;113:202–208. doi: 10.1046/j.1365-2141.2001.02705.x. [DOI] [PubMed] [Google Scholar]
- Cullen MH, Lister TA, Brearley RI, Shand WS, Stansfeld AG. Histological transformation of non-Hodgkin’s lymphoma: a prospective study. Cancer. 1979;44:645–651. doi: 10.1002/1097-0142(197908)44:2<645::aid-cncr2820440234>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Dahia PL, Aguiar RC, Honegger J, Fahlbush R, Jordan S, Lowe DG, Lu X, Clayton RN, Besser GM, Grossman AB. Mutation and expression analysis of the p27/kip1 gene in corticotrophin-secreting tumours. Oncogene. 1998;16:69–76. doi: 10.1038/sj.onc.1201516. [DOI] [PubMed] [Google Scholar]
- Davies AJ, Lee AM, Taylor C, Clear AJ, Goff LK, Iqbal S, Cuthbert-Heavens D, Calaminici M, Norton AJ, Lister TA, Fitzgibbon J. A limited role for TP53 mutation in the transformation of follicular lymphoma to diffuse large B-cell lymphoma. Leukemia. 2005;19:1459–1465. doi: 10.1038/sj.leu.2403802. [DOI] [PubMed] [Google Scholar]
- Elenitoba-Johnson KS, Gascoyne RD, Lim MS, Chhanabai M, Jaffe ES, Raffeld M. Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood. 1998;91:4677–4685. [PubMed] [Google Scholar]
- Elenitoba-Johnson KS, Jenson SD, Abbott RT, Palais RA, Bohling SD, Lin Z, Tripp S, Shami PJ, Wang LY, Coupland RW, Buckstein R, Perez-Ordonez B, Perkins SL, Dube ID, Lim MS. Involvement of multiple signaling pathways in follicular lymphoma transformation: p38-mitogen-activated protein kinase as a target for therapy. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7259–7264. doi: 10.1073/pnas.1137463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin AJ, Simon RM, Osborne CK, Merrill J, Young RC, Berard CW. An autopsy study of histologic progression in non-Hodgkin’s lymphomas. 192 cases from the National Cancer Institute. Cancer. 1983;52:393–398. doi: 10.1002/1097-0142(19830801)52:3<393::aid-cncr2820520302>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature Reviews. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Geschwind JF, Georgiades CS, Ko YH, Pedersen PL. Recently elucidated energy catabolism pathways provide opportunities for novel treatments in hepatocellular carcinoma. Expert Review of Anticancer Therapy. 2004;4:449–457. doi: 10.1586/14737140.4.3.449. [DOI] [PubMed] [Google Scholar]
- Glas AM, Kersten MJ, Delahaye LJ, Witteveen AT, Kibbelaar RE, Velds A, Wessels LF, Joosten P, Kerkhoven RM, Bernards R, van Krieken JH, Kluin PM, van’t Veer LJ, de Jong D. Gene expression profiling in follicular lymphoma to assess clinical aggressiveness and to guide the choice of treatment. Blood. 2005;105:301–307. doi: 10.1182/blood-2004-06-2298. [DOI] [PubMed] [Google Scholar]
- Goff LK, Neat MJ, Crawley CR, Jones L, Jones E, Lister TA, Gupta RK. The use of real-time quantitative polymerase chain reaction and comparative genomic hybridization to identify amplification of the REL gene in follicular lymphoma. British Journal of Haematology. 2000;111:618–625. doi: 10.1046/j.1365-2141.2000.02352.x. [DOI] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Horning SJ, Rosenberg SA. The natural history of initially untreated low-grade non-Hodgkin’s lymphomas. The New England Journal of Medicine. 1984;311:1471–1475. doi: 10.1056/NEJM198412063112303. [DOI] [PubMed] [Google Scholar]
- Hough RE, Goepel JR, Alcock HE, Hancock BW, Lorigan PC, Hammond DW. Copy number gain at 12q12–14 may be important in the transformation from follicular lymphoma to diffuse large B-cell lymphoma. British Journal of Cancer. 2001;84:499–503. doi: 10.1054/bjoc.2000.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SM, Chabner BA, DeVita VT, Jr, Simon R, Berard CW, Jones RB, Garvin AJ, Canellos GP, Osborne CK, Young RC. Histologic progression in non-Hodgkin’s lymphoma. Blood. 1982;59:258–264. [PubMed] [Google Scholar]
- Iqbal S, Jenner MJ, Summers KE, Davies AJ, Matthews J, Norton AJ, Calaminici M, Rohatiner AZ, Fitzgibbon J, Lister TA, Goff LK. Reliable detection of clonal IgH/Bcl2 MBR rearrangement in follicular lymphoma: methodology and clinical significance. British Journal of Haematology. 2004a;124:325– 328. doi: 10.1046/j.1365-2141.2003.04796.x. [DOI] [PubMed] [Google Scholar]
- Iqbal J, Sanger WG, Horsman DE, Rosenwald A, Pickering DL, Dave B, Dave S, Xiao L, Cao K, Zhu Q, Sherman S, Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Ott G, Muller-Hermelink HK, Delabie J, Braziel RM, Jaffe ES, Campo E, Lynch JC, Connors JM, Vose JM, Armitage JO, Grogan TM, Staudt LM, Chan WC. BCL2 translocation defines a unique tumor subset within the germinal center B-cell- like diffuse large B-cell lymphoma. The American Journal of Pathology. 2004b;165:159–166. doi: 10.1016/s0002-9440(10)63284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53- regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- Labuhn M, Jones G, Speel EJ, Maier D, Zweifel C, Gratzl O, Van Meir EG, Hegi ME, Merlo A. Quantitative real-time PCR does not show selective targeting of p14(ARF) but concomitant inactivation of both p16(INK4A) and p14(ARF) in 105 human primary gliomas. Oncogene. 2001;20:1103–1109. doi: 10.1038/sj.onc.1204197. [DOI] [PubMed] [Google Scholar]
- Leonard BM, Hetu F, Busque L, Gyger M, Belanger R, Perreault C, Roy DC. Lymphoma cell burden in progenitor cell grafts measured by competitive polymerase chain reaction: less than one log difference between bone marrow and peripheral blood sources. Blood. 1998;91:331–339. [PubMed] [Google Scholar]
- Lo Coco F, Gaidano G, Louie DC, Offit K, Chaganti RS, Dalla-Favera R. p53 mutations are associated with histologic transformation of follicular lymphoma. Blood. 1993;82:2289–2295. [PubMed] [Google Scholar]
- Lossos IS, Levy R. Higher-grade transformation of follicle center lymphoma is associated with somatic mutation of the 5′ noncoding regulatory region of the BCL-6 gene. Blood. 2000;96:635–639. [PubMed] [Google Scholar]
- Lossos IS, Alizadeh AA, Diehn M, Warnke R, Thorstenson Y, Oefner PJ, Brown PO, Botstein D, Levy R. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8886–8891. doi: 10.1073/pnas.132253599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Climent JA, Alizadeh AA, Segraves R, Blesa D, Rubio-Moscardo F, Albertson DG, Garcia-Conde J, Dyer MJ, Levy R, Pinkel D, Lossos IS. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003;101:3109–3117. doi: 10.1182/blood-2002-07-2119. [DOI] [PubMed] [Google Scholar]
- Marutani M, Tonoki H, Tada M, Takahashi M, Kashiwazaki H, Hida Y, Hamada J, Asaka M, Moriuchi T. Dominant-negative mutations of the tumor suppressor p53 relating to early onset of glioblastoma multiforme. Cancer Research. 1999;59:4765–4769. [PubMed] [Google Scholar]
- Matolcsy A, Warnke RA, Knowles DM. Somatic mutations of the translocated bcl-2 gene are associated with morphologic transformation of follicular lymphoma to diffuse large-cell lymphoma. Annals of Oncology. 1997;8(Suppl 2):119–122. [PubMed] [Google Scholar]
- Nagy M, Balazs M, Adam Z, Petko Z, Timar B, Szereday Z, Laszlo T, Warnke RA, Matolcsy A. Genetic instability is associated with histological transformation of follicle center lymphoma. Leukemia. 2000;14:2142–2148. doi: 10.1038/sj.leu.2401978. [DOI] [PubMed] [Google Scholar]
- Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. The Journal of Biological Chemistry. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- Price CG, Meerabux J, Murtagh S, Cotter FE, Rohatiner AZ, Young BD, Lister TA. The significance of circulating cells carrying t(14;18) in long remission from follicular lymphoma. Journal of Clinical Oncology. 1991;9:1527–1532. doi: 10.1200/JCO.1991.9.9.1527. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large B-cell lymphoma. The New England Journal of Medicine. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Sander CA, Yano T, Clark HM, Harris C, Longo DL, Jaffe ES, Raffeld M. p53 mutation is associated with progression in follicular lymphomas. Blood. 1993;82:1994–2004. [PubMed] [Google Scholar]
- Schoder H, Noy A, Gonen M, Weng L, Green D, Erdi YE, Larson SM, Yeung HW. Intensity of 18fluorodeoxyglucose uptake in positron emission tomography distinguishes between indolent and aggressive non-Hodgkin’s lymphoma. Journal of Clinical Oncology. 2005;23:4643–4651. doi: 10.1200/JCO.2005.12.072. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, Staudt LM. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- Shi XB, Nesslinger NJ, Deitch AD, Gumerlock PH, de Vere White RW. Complex functions of mutant p53 alleles from human prostate cancer. The Prostate. 2002;51:59–72. doi: 10.1002/pros.10072. [DOI] [PubMed] [Google Scholar]
- Szereday Z, Csernus B, Nagy M, Laszlo T, Warnke RA, Matolcsy A. Somatic mutation of the 5′ noncoding region of the BCL-6 gene is associated with intraclonal diversity and clonal selection in histological transformation of follicular lymphoma. The American Journal of Pathology. 2000;156:1017–1024. doi: 10.1016/S0002-9440(10)64969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos S, Hofmann WK, Grogan TM, Krug U, Schrage M, Miller TP, Braun JG, Wachsman W, Koeffler HP, Said JW. Gene expression profile of serial samples of transformed B-cell lymphomas. Laboratory Investigation. 2003;83:271–285. doi: 10.1097/01.lab.0000053913.85892.e9. [DOI] [PubMed] [Google Scholar]
- Williams CD, Harrison CN, Lister TA, Norton AJ, Blystad AK, Coiffier B, Taghipour G, Schmitz N, Goldstone AH. High-dose therapy and autologous stem-cell support for chemosensitive transformed low-grade follicular non-Hodgkin’s lymphoma: a case-matched study from the European Bone Marrow Transplant Registry. Journal of Clinical Oncology. 2001;19:727–735. doi: 10.1200/JCO.2001.19.3.727. [DOI] [PubMed] [Google Scholar]
- Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Jaffe ES, Longo DL, Raffeld M. MYC rearrangements in histologically progressed follicular lymphomas. Blood. 1992;80:758–767. [PubMed] [Google Scholar]


