Fig 2.
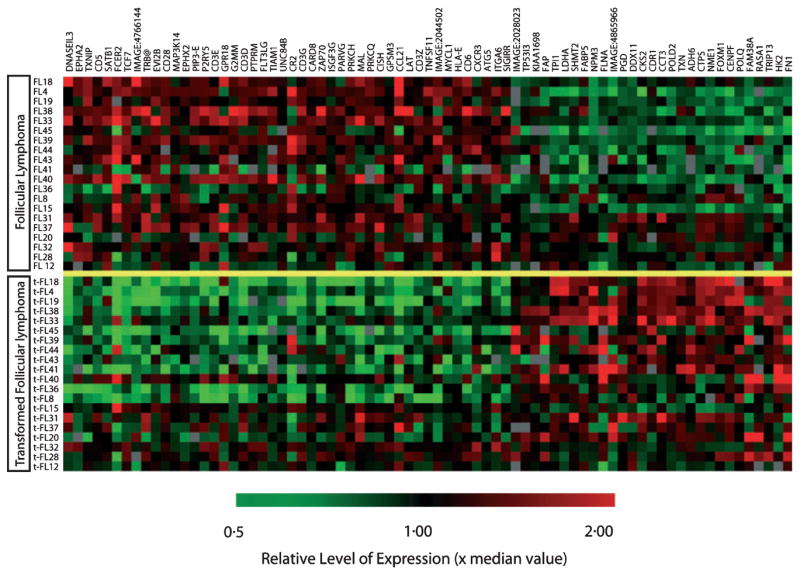
Differential gene expression in supervised analysis between paired follicular lymphoma and transformed follicular lymphoma (t-FL) samples. From 118 clones, 76 genes represented by distinct UniGene clusters were highly significant with a P-value of <0.001 between different histological phenotypes. The gene identification is annotated to the right. Genes involved in cellular proliferation (POLQ, POLD2, CENPF, CTPS, NME1, FOXM11, DDX11, SHMT2, TXN, CCT3, TPI1 and NPM3) predominated amongst those up regulated upon transformation. In addition, genes involved in metabolism (HK2, PGD, FABP5 and LDHA), cytoskeletal components (FN1 and FLNB), molecular chaperones (CCT and NPM3) and apoptotic genes (TXN, TP53I3 and HK2) were also up regulated in t-FL. T cell associated (CD3E, CD3G, CD6, CD28, CXCR3, LAT, TRB@, PRKCQ, MAL, ZAP70, FLT3LG, TCF7, G2MM and DNASEIL3) and follicular dendritic cell genes (CR2 and FCER2) predominated amongst genes that demonstrated significantly reduced expressions upon transformation.
