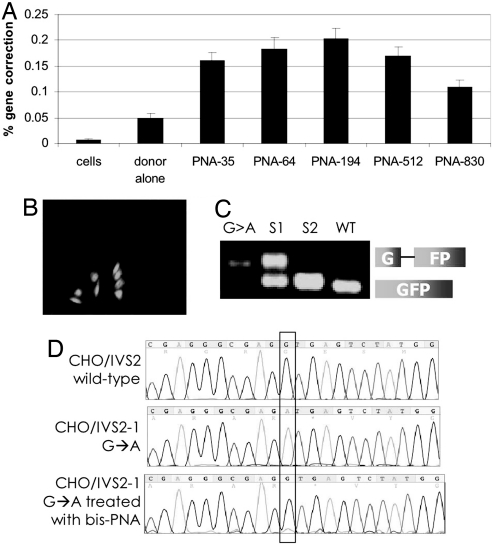
Fig. 2.
bis-PNAs induce correction of the IVS2–1 mutation by donor DNAs in the CHO-GFP/IVS2–1G→A reporter cell assay. (A) Gene correction was measured as the percentage of transfected cells expressing GFP by FACS analysis. CHO-GFP/IVS2–1G→A cells, synchronized in S phase, were transfected with no oligonucleotides (cells), with IVS2 donor DNA (donor alone), or with the indicated bis-PNA plus IVS2 donor DNA. Bar graph represents data collected from at least three independent experiments, and the error bars indicate standard errors. (B) Fluorescence micrograph of PNA and donor DNA-treated cells 48 h after transfection showing GFP-expressing CHO-GFP/IVS2–1G→A cells in a field of predominantly GFP-negative, uncorrected cells. (C) RT-PCR analysis of RNA harvested from GFP-positive CHO-GFP/IVS2–1G→A cells that were isolated by FACS. The IVS2–1 splicing mutation yields a longer RT-PCR product. G > A: untreated CHO-GFP/IVS2–1G→A cells; S1 and S2: first and second sort, respectively, of CHO-GFP/IVS2–1G→A cells treated with bis-PNA-35 and IVS2 donor DNA; WT: CHO-GFP/IVS2wt cells with wild-type and thus properly spliced transcript. Sorted CHO-GFP/IVSG→A cells that were treated with bis-PNA and IVS2 donor DNA show increasing proportions of RT-PCR product matching that of CHO-GFP/IVS2wt cells, indicating restoration of correct splicing. (D) Genomic DNA from sorted GFP-expressing cells maintained in culture for 1 month was harvested and sequenced to verify genomic modification at the target site. Chromograms are from untreated CHO-GFP/IVS2wt cells containing the wild-type intron (Top), untreated CHO-GFP/IVS2–1G→A cells with the IVS2–1 G→A mutation (Middle), and CHO-GFP/IVS2–1G→A cells treated with bis-PNA-35 and IVS2 donor DNAs, and sorted by FACS for GFP-positive cells (Bottom).