Figure 1.
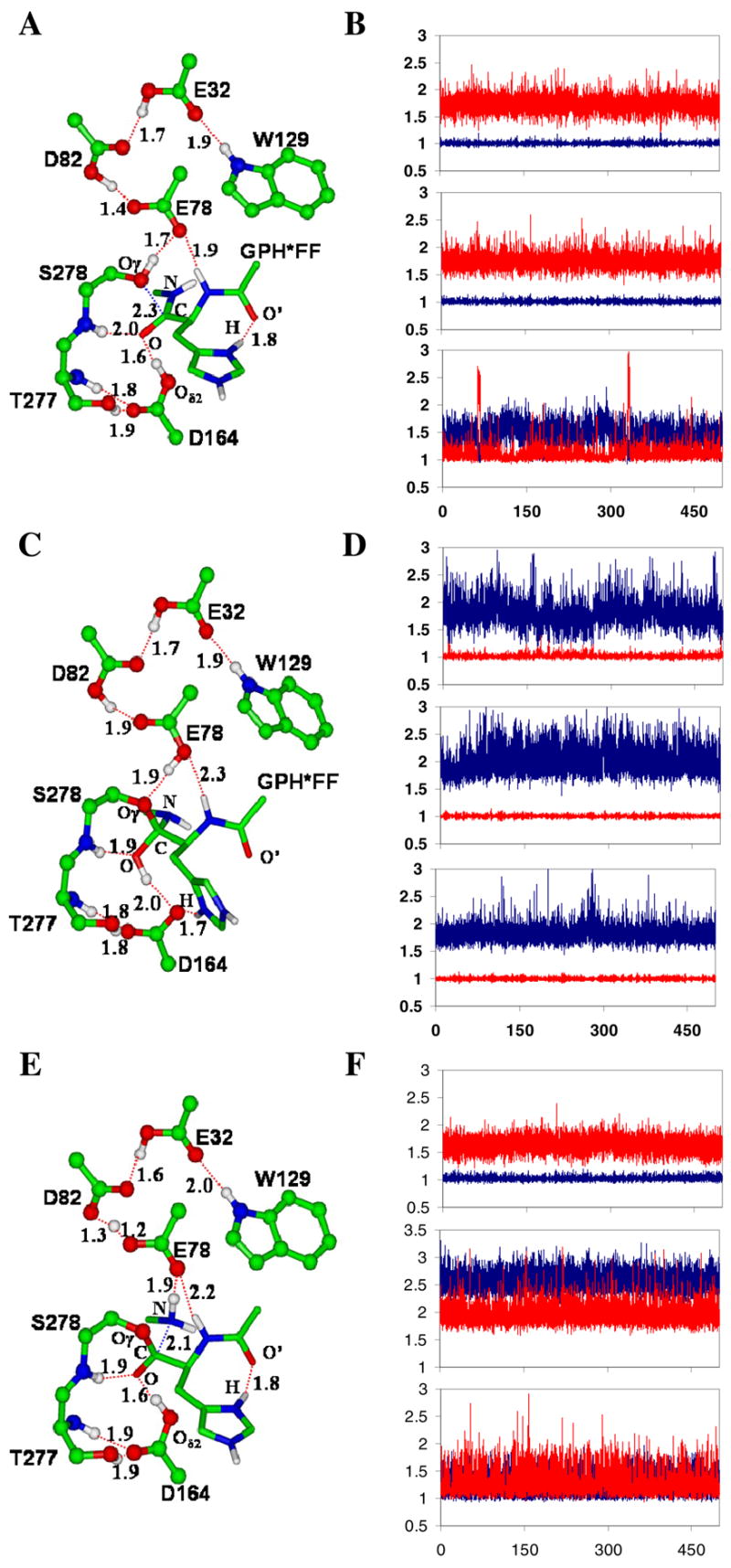
The average active-site structures of the kumamolsin-As-substrate (GPH*FF) complex, tetrahedral intermediate and acyl-enzyme obtained from the QM/MM MD simulations and the motions of the three key protons involved in the catalysis. The initial structures of the simulations for the tetrahedral intermediate and acyl-enzyme were based on the structures from the free energy simulations (see below). (A) The average structure of the substrate complex. The enzyme is plotted in ball and stick and the substrate in stick. Hydrogen bonds (H-bonds) are indicated by red dashed lines. Only the sidechain of the P1 (His) residue of the substrate is shown for clarity. The catalytic triad contains the Ser278, Glu78 and Asp82 residues. The His sidechain at P1 donates an H-bond to the C=O′ group of Pro at P2, in addition to the carboxylate of Asp179 (not shown). The backbone N-H group of the P1 residue donates a hydrogen bond to Glu78. (B) Fluctuations of the distances (in Å) related to proton motions (vibrations) between Asp164 and substrate, Ser278 and Glu78, and Glu78 and Asp82 in the substrate complex. Top panel: fluctuations of the H-O(D164) and H…O(substrate) distances as function of time. Red, r[H…O(substrate)]; blue, r[H-O(D164)]. The proton is on Asp 164 and is involved in an H-bond with the C=O group of the substrate (see also Figure 1A). Middle panel: the fluctuations of the H-O(S278) and H…O(E78) distances as function of time. Red, r[H…O(E78)]; blue, r[H-O(S278)]. The proton is on Ser278 and is involved in an H-bond with Glu78. Bottom panel: the fluctuations of the H…O(E78) and H-O(D82) distances as function of time. Red, r[H-O(D82)]; blue, r[H…O(E78)]. The H-bond formed between Glu78 and Asp82 has a short distance of !1.5 Å, and the proton is mainly located on Asp82. (C) The average structure of the tetrahedral intermediate after the nucleophilic attack by Ser278 on the substrate. The His sidechain at P1 now interacts with Oδ2 of Asp164 and may, therefore, play a role in stabilizing the charge formation on Asp164. The H-bond between the backbone N-H group of the P1 residue and Glu78 becomes significantly weaker due to the protonation of Glu78. (D) Fluctuations of the distances related to proton motions (vibrations) in the tetrahedral intermediate complex. Top panel: the fluctuations of the H…O(D164) and H-O(tetrahedral intermediate) distances as functions of time. Red, r[H-O(tetrahedral intermediate)]; blue, r[H…O(D164)]. The proton is on the tetrahedral intermediate and is involved in an H-bond with Asp164 (see Figure 1C). Middle panel: the fluctuations of the H…O(S278) and H-O(E78) distances as function of time. Red, r[H-O(E78)]; blue, r[H…O(S278)]. The proton is on Glu78 and is involved in an H-bond with Ser278. Bottom panel: the fluctuations of the H…O(E78) and H-O(D82) distances as function of time. Red, r[H-O(D82)]; blue, r[H…O(E78)]. (E) The average structure of the acyl-enzyme with the C-N bond broken. (F) Fluctuations of the distances related to proton motions (vibrations) in the acy-enzyme. Top panel: the fluctuations of the H-O(D164) and H…O distances as function of time. Red, r[H…O]; blue, r[H-O(D164)]. Middle panel: the fluctuations of the H…O(S278) and H…O(E78) distances as function of time. Red, r[H…O(E78)]; blue, r[H…O(S278)]. Bottom panel: the fluctuations of the H…O(E78) and H…O(D82) distances as function of time. Red, r[H…O(D82)]; blue, r[H…O(E78)].
