Figure 3.
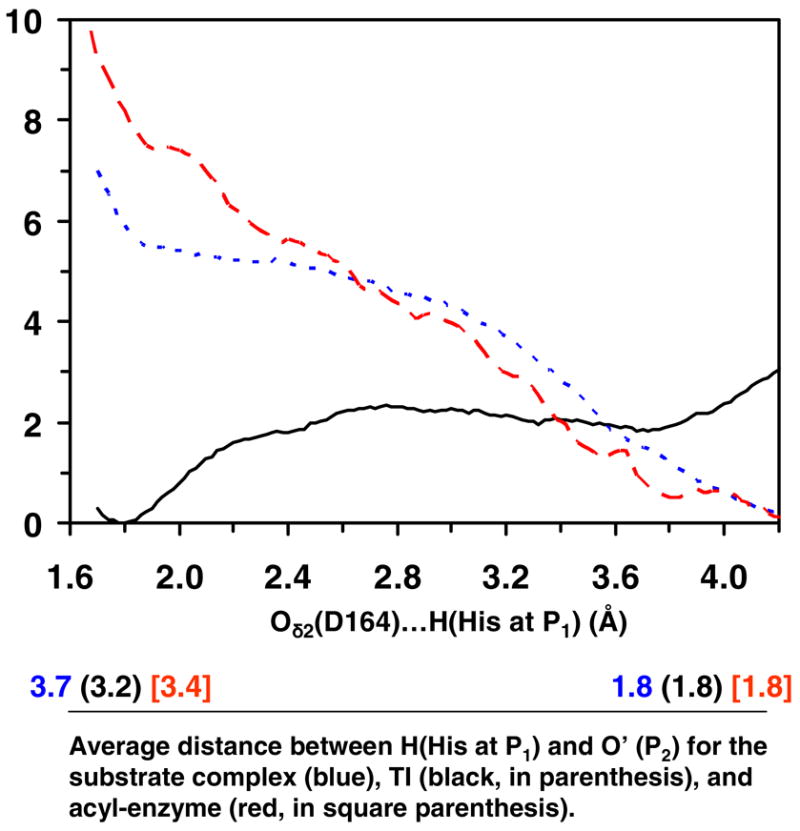
The free energy change as the His residue at P1 moves between Oδ2 of Asp164 and O′ of the C=O′ group of Pro at P2 in the substrate complex, tetrahedral intermediate and acyl-enzyme for kumamolisin-As (see Figures 1A, 1C and 1E). The reaction coordinate is the distance between the hydrogen atom (H) of the His residue at P1 and Oδ2 of Asp164. The corresponding average distances between H and O′ are given below. For instance, when the reaction coordinate is 4.2 Å for the substrate complex, the average distance for R(H…O′) is about 1.8 Å based on the trajectory of the corresponding window (i.e., H forms a hydrogen bond with the C=O′ group of Pro at P2). Blue dotted line: the free energy change in the substrate complex. For the substrate complex, the conformations for which His at PI forms a hydrogen bond with the C=O′ group of Pro at P2 (i.e., the structure shown in Figure 1A) are considerable more stable (by about 5–6 kcal/mol) than the conformations for which the His residue interacts with Oδ2 of Asp164. Black solid line: the free energy change in the tetrahedral intermediate. For the tetrahedral intermediate, the conformations for which the His at P1 forms a hydrogen bond with Oδ2 of Asp164 (i.e., the structure shown in Figure 1C) are more stable (by about 2 kcal/mol) than the conformations for which His interacts with C=O group of Pro at P2. Red dashed line: the free energy change in the acyl-enzyme. The conformations for which the His residue forms a hydrogen bond with the C=O′ group of Pro at P2 (i.e., the structure shown in Figure 1E) are considerable more stable than the conformations for which His at P1 interacts with Oδ2 of Asp164.
