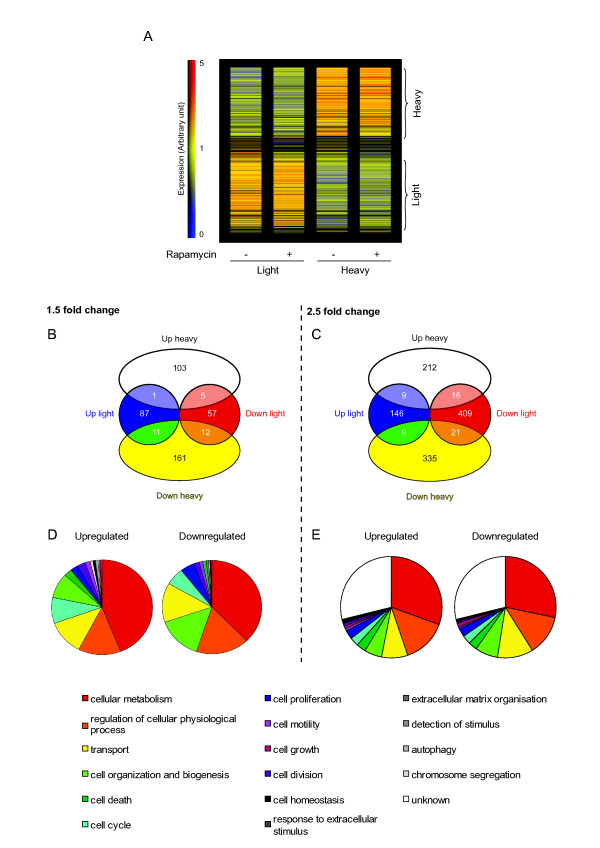
Figure 2.
Microarray analysis. (A). Hierarchical clustering of relative expression. Each column represents the different conditions. On the right the vertical bar indicates probe set intensity values (indicated as Expression) in arbitrary units. The zero value indicates absence, with blue indicating a low level and red a high level of expression (the maximum value being fixed as 5). The vertical brackets on the right indicate that transcripts have been grouped into those over-represented in the light polysomes (lower) or in the heavy polysomes (upper). (B). After removal of probe set intensity values < 100, and the application of a ×1.5 fold cut-off, the regulated genes were classified into four groups. The values indicate the number of transcripts in each group. (C). In a second approach, a ×2.5 fold cut-off was applied. (D) and (E). Functional classification using Gene Ontology of the genes either up-regulated or down-regulated (as depicted in panel b and in Additional File 1, and panel c and in Additional File 2, respectively). The unknown fraction represents genes not annotated in the Gene Ontology database.