Abstract
Mycobacterium tuberculosis adapts to the environment by selecting for advantageous single-nucleotide polymorphisms (SNPs). We studied whether advantageous SNPs could be distinguished from neutral mutations within genes associated with drug resistance. A total of 1,003 clinical isolates of M. tuberculosis were related phylogenetically and tested for the distribution of SNPs in putative drug resistance genes. Drug resistance-associated versus non-drug-resistance-associated SNPs in putative drug resistance genes were compared for associations with single versus multiple-branch outcomes using the chi-square and Fisher exact tests. All 286 (100%) isolates containing isoniazid (INH) resistance-associated SNPs had multibranch distributions, suggestive of multiple ancestry and convergent evolution. In contrast, all 327 (100%) isolates containing non-drug-resistance-associated SNPs were monophyletic and thus showed no evidence of convergent evolution (P < 0.001). Convergence testing was then applied to SNPs at position 481 of the iniA (Rv0342) gene and position 306 of the embB gene, both potential drug resistance targets for INH and/or ethambutol. Mutant embB306 alleles showed multibranch distributions, suggestive of convergent evolution; however, all 44 iniA(H481Q) mutations were monophyletic. In conclusion, this study validates convergence analysis as a tool for identifying mutations that cause INH resistance and explores mutations in other genes. Our results suggest that embB306 mutations are likely to confer drug resistance, while iniA(H481Q) mutations are not. This approach may be applied on a genome-wide scale to identify SNPs that impact antibiotic resistance and other types of biological fitness.
Mycobacterium tuberculosis continuously adapts to the environment by selecting for single-nucleotide polymorphism (SNPs) that can confer selective advantages on the bacterial populations. While this is most apparent in the development of drug resistance (29, 35, 37, 38), similar events may be related to the increased fitness and virulence observed for some clinical strains. It would be of great interest to assign a role to each of the numerous SNPs that have been discovered among clinical isolates of M. tuberculosis. M. tuberculosis would appear to be an ideal bacterial species for this type of analysis. M. tuberculosis genomes are highly conserved and do not naturally contain plasmids or exhibit significant levels of horizontal gene transfer (11, 14). Only 1,075 SNPs and 86 large-sequence polymorphisms have been detected between the complete genome sequences of the laboratory strain H37Rv and the clinical strain CDC1551 (13). M. tuberculosis also exhibits considerable differences between strains in growth rates, the ability to acquire drug resistance, and other virulence attributes (3, 10, 15, 22, 24, 25, 36), despite this relative lack of genetic polymorphism. Finally, 10 M. tuberculosis genomes have now been completely sequenced (9, 13; Broad Institute, Mycobacterium tuberculosis Database [http://www.broad.mit.edu/annotation/genome/mycobacterium_tuberculosis_spp/MultiHome.html]), providing excellent opportunities to study the genetics of phenotypic variation using a comparative genomic approach.
Unfortunately, even the relatively small numbers of polymorphisms that exist among clinical M. tuberculosis strains loom large when considered in light of the allelic exchange experiments and subsequent bioassays that must be performed in order to definitively determine the phenotypic relevance of a particular mutation. As a result, in-depth studies of genetic changes among clinical isolates have been restricted to the few drug resistance-associated mutations where the evidence for a true role in disease phenotype is already strong. For example, katG(S315T) (20, 27, 32, 41), inhA −15 C-to-T, and inhA(S94A) mutations (40) have been proven to confer resistance to the drug isoniazid (INH) in careful allelic exchange studies, while some drug resistance-associated mutations originally believed to confer resistance to INH [i.e., mutations kasA(G269S) and kasA(G312S)] have been shown not to be involved in INH resistance by using a similar approach (40). The complexity of these studies demonstrates the critical need to complement comparative genomic investigations with simpler methods that can identify the mutations most likely to affect bacterial phenotypes such as drug resistance or pathogenicity.
Phylogenetic analysis has the potential to aid in studies of bacterial pathogenesis by ordering isolates into genetically related groups and by situating isolates and genetic polymorphisms with potential biological relevance within an evolutionary context (2, 11, 14). We have hypothesized that mutations conferring a selective advantage on a bacterial species can also be identified using phylogenetic techniques (2). Neutral mutations are likely to appear as unique events in a finite study population of a species such as M. tuberculosis. This is because mutations occur so infrequently that they are unlikely to be detected unless first amplified by selective pressures against wild-type bacteria. Random neutral mutations will become detectable only if they become linked to an advantageous change within the bacterial population. However, strictly neutral mutations are unlikely to become independently linked to advantageous mutations a second time, and without a selective advantage, most of these mutations will be lost by random drift and bottlenecks. Phylogenetic analysis can be used to confirm that a mutation arose only once in a population and to identify such a mutation as likely to be neutral. Conversely, mutations that provide a selective advantage to M. tuberculosis should increase in frequency and become detectable each time they arise. This suggests that the independent presence of a specific polymorphism on multiple phylogenetic branches would be strong evidence for convergent evolution and biological relevance.
Several sets of synonymous SNPs have been described that enable relatively detailed phylogenetic analysis of M. tuberculosis (2, 4, 11, 14). More recently, nine synonymous SNPs were described using cluster analysis that can be used in place of a much larger number of SNPs to divide the M. tuberculosis species into 10 distinct groups and subgroups that have been termed SNP cluster groups (SCGs) and subgroups (SCG subgroups) 1, 2, 3a, 3b, 3c, 4, 5, 6a, 6b, and 7 (1). More than 1,000 M. tuberculosis isolates of clinical origin have also been analyzed recently for mutations hypothesized to contribute to resistance to the antibiotics INH and ethambutol (EMB) (17, 18). Although many of these mutations were uniquely associated with drug-resistant strains (and likely to provide a selective advantage to the bacterium), a number of others were found to be present in both drug-susceptible and drug-resistant isolates (and likely to be neutral polymorphisms with respect to resistance). Here, we examined this large collection of well-characterized M. tuberculosis isolates in order to formally test the hypothesis that phylogenetic analysis for convergent evolution can identify mutations that provide a strong selective advantage to M. tuberculosis. We further explore the phylogenetics of an additional mutation in the iniA gene (Rv0341) with uncertain significance. This analysis revealed for the first time mutations that are likely to be beneficial to M. tuberculosis versus mutations that are unlikely to provide a selective advantage in antibiotic environments. This study establishes the utility of phylogenetic investigations in determining the biological relevance of mutations discovered through comparative genomics and suggests that phylogenetic investigations may be applied more broadly in future genomic studies.
MATERIALS AND METHODS
M. tuberculosis clinical isolates.
A total of 1,013 clinical M. tuberculosis isolates were obtained from reference laboratories or major medical centers in Australia, Colombia, India, Mexico, New York City, Spain, and Texas (17, 18). Of the 1,013 isolates, 10 were unavailable to this study because the DNA was no longer available or had degraded, leaving a total of 1,003 isolates included in the analysis.
Drug susceptibility testing and strain typing.
All isolates were subjected to susceptibility testing and DNA fingerprinting as described previously (18). In brief, each center or laboratory tested susceptibility to at least INH, rifampin (RIF), streptomycin (STR), and EMB by the agar proportion method (21) (Colombia, India, New York, and Spain), the Bactec MGIT 960 system (19) (Australia), or the radiometric Bactec 460 method (23) (Mexico and Texas). DNA fingerprinting, performed as described previously (18), demonstrated that this study population included 339 unique and 267 clustered drug-susceptible isolates, 190 unique and 199 clustered isolates resistant to INH, and 47 unique and 55 clustered isolates resistant to EMB (this total is greater than the size of the total study because 43 unique and 54 clustered EMB-resistant isolates were also INH resistant). The restriction fragment length polymorphism patterns for three of the isolates were unknown.
Detection of mutations in clinical isolates of M. tuberculosis.
INH resistance-associated mutations in the katG, kasA, mabA, inhA, oxyR, ahpC, and ndh genes (or upstream promoter regions) and EMB resistance-associated mutations in embB codon 306 were detected as described previously (16, 18). Mutations at positions 481 of iniA and 95 of gyrA were detected using hairpin-shaped primer (HP) assays designed for this study (Table 1) and performed as described previously (16). All mutations were confirmed by DNA sequencing, except when they were abundant, in which case a subset of mutations was confirmed. iniA481 HP assays with uncertain results were retested by a molecular beacon assay (Table 1), and subsets of the molecular beacon assays were confirmed by DNA sequencing. All mutations occurring in at least three different isolates were included in this study. Two INH resistance-associated mutations [katG(N138D), resulting from an aac→Gac change, and katG(A172T), resulting from a gcg→Acg change] and one non-resistance-associated mutation [katG(G316S), resulting from a ggc→Agc change] were present in only two isolates and mapped to a single phylogenetic branch. (Capital letters in codons indicate mutated nucleotides.) These three mutations were excluded from analysis because their frequencies were deemed to be too low to permit reliable detection of multibranch distributions.
TABLE 1.
Molecular beacon and HP assays used for the first time in this study
| Assay | Purpose | Primer or MBa | Sequenceb |
|---|---|---|---|
| IniA(H481Q) (cat to caG) | MB assay forward primer | FiniA481 | ggccggatggaatcgaaa |
| MB assay reverse primer | RiniA481 | cgccgccataggaaccc | |
| MB complementary to iniA(481H) | MBIniA481T | FAM-TCCGCGcggggccataaaatgat CGCGGA- DABCYL | |
| MB complementary to iniA(481Q) | MBIniA481G | TET-ACCGCCcggggccagaaaatgat GGCGGT- DABCYL | |
| HP assay primer complementary to iniA(481H) | FHPIniA481 | atggccactgccccggggccat | |
| HP assay primer complementary to iniA(481Q) | FHPIniAH481Q | ctggccactgccccggggccag | |
| HP assay shared primer | RIniA481 | cgccgccataggaaccc | |
| GyrA(T95S) (acc to aGc) | HP assay primer complementary to gyrA95 T | RHPGyrA95 | ccctgggccatccgcaccaggg |
| HP assay primer complementary to gyrA95 S | RHPGyrAT95S | gcctgggccatccgcaccaggc | |
| HP assay forward primer | FGyrA95 | cgagaccatgggcaactaccacc |
MB, molecular beacon.
Capital letters indicate molecular beacon stem sequences; lowercase letters indicate the probe region. FAM, 6-carboxyfluorescein; DABCYL, 4{[4′-(dimethylamino)phenyl]azo}benzoic acid; TET, tetrachloro-6-carboxy fluorescein.
Phylogenetic and statistical analysis.
Each isolate (n = 1,003) was assigned to an SCG as described previously (1) and then mapped onto the previously described phylogenetic tree (11) using nine SNP markers developed for this purpose (1). These SNP markers are prevalent in different proportions across the various SCGs, and their distribution in the tree diagram was observed; the presence of a marker in more than one branch of the tree was noted. The SNP type of the INH-resistant isolates had been reported previously (6). The chi-square and Fisher exact tests were used to assess association in single versus multiple-branch outcomes and drug-resistant versus non-drug-resistant status.
RESULTS
SCG assignments.
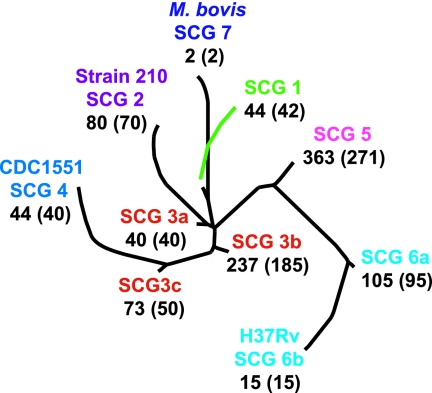
The 1,003 M. tuberculosis isolates in this study were SNP typed, assigned to 1 of 10 SCGs and SCG subgroups, and situated on a previously described phylogenetic tree (11) using nine SNP markers developed for this purpose (1). The number of isolates ranged from 15 in SCG 6b to 363 in SCG 5 based on the phylogenetic classification (Fig. 1). Strains of all seven SCGs were originally isolated in Mexico, and only strains of three SCGs were originally isolated in Spain (Table 2). Three SCGs were recovered from patients in all six countries of origin (seven populations); four were recovered from four to five countries of origin; and three were recovered from fewer than four countries, presumably because of smaller sample sizes (Table 2). Isolates belonging to all 10 SCGs and subgroups were present, although a larger proportion of isolates belonged to SCGs 3b and 5. Two isolates belonged to SCG 7, which harbors Mycobacterium bovis. One of these isolates was obtained from a patient with AIDS who developed a tuberculosis-like disease after receiving an M. bovis BCG vaccination. No additional information was available about the second SCG-7 isolate. All of the isolates had been characterized for a large number of mutations associated with resistance to INH or EMB in previous studies (17, 18). In the current study, isolates were also tested for mutations at iniA481, which has been postulated to be associated with INH or EMB resistance in clinical M. tuberculosis isolates (28, 29), and gyrA95, which is in the quinolone resistance-determining region but has been shown not to be associated with resistance to quinolones (34).
FIG. 1.
Phylogenetic locations of the study isolates. Study isolates were placed on a previously created phylogenetic tree (which had been based on an analysis of 212 SNPs in a global collection of 327 M. tuberculosis isolates). The SCGs, including subgroups, and the positions of the M. tuberculosis reference strains 210, CDC1551, and H37Rv and of M. bovis strain AF2122/97 are indicated. The total number of distinct strains, as defined by DNA fingerprinting, within each SCG is given in parentheses.
TABLE 2.
Distribution of the SCGs of the isolates studied by geographic origin
| Geographic origin | No. of isolates | No. (%) of isolates in the following SCG:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3a | 3b | 3c | 4 | 5 | 6a | 6b | 7 | ||
| Australia | 56 | 19 (34) | 17 (30) | 6 (11) | 4 (7) | 0 | 1 (2) | 5 (9) | 2 (4) | 2 (4) | 0 |
| Colombia | 301 | 1 (0) | 0 | 2 (1) | 88 (29) | 9 (3) | 7 (2) | 170 (56) | 23 (8) | 1 (0) | 0 |
| India | 39 | 4 (10) | 1 (3) | 28 (72) | 1 (3) | 0 | 0 | 4 (10) | 1 (3) | 0 | 0 |
| Mexico | 196 | 2 (1) | 1 (1) | 1 (1) | 76 (39) | 19 (10) | 20 (10) | 45 (23) | 24 (12) | 6 (3) | 2 (1) |
| New York City | 146 | 10 (7) | 23 (16) | 1 (1) | 27 (18) | 27 (18) | 5 (3) | 39 (27) | 11 (8) | 3 (2) | 0 |
| Spain | 109 | 0 | 0 | 0 | 25 (23) | 0 | 0 | 69 (63) | 15 (14) | 0 | 0 |
| Texas | 156 | 8 (5) | 38 (24) | 2 (1) | 16 (10) | 18 (12) | 11 (7) | 31 (20) | 29 (19) | 3 (2) | 0 |
| Total isolates | 1,003 | 44 (4) | 80 (8) | 40 (4) | 237 (24) | 73 (7) | 44 (4) | 363 (36) | 105 (11) | 15 (1) | 2 (0) |
| Total strainsa | 810 | 42 (5) | 70 (9) | 40 (5) | 185 (23) | 50 (6) | 40 (5) | 271 (33) | 95 (12) | 15 (2) | 2 (0) |
“Total strains” refers to the number of isolates with different restriction fragment length polymorphism patterns, mutation profiles, and SCGs.
INH-resistant SNPs.
The phylogenetic distribution of “INH-resistant SNPs” was examined. These SNPs had been identified previously to be present in INH-resistant M. tuberculosis isolates but not in INH-susceptible isolates (18). INH-resistant SNPs included mutations in katG, the inhA promoter, the inhA open reading frame, and the ahpC promoter except for aphC −46. The combined results subdivided by SCGs are shown in the supplemental material.
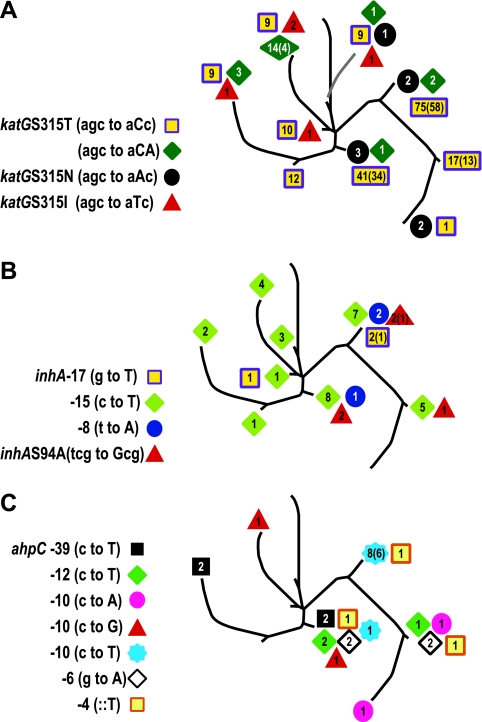
When plotted on the phylogenetic tree, the katG SNP alleles were found to be distributed across multiple branches. Each branch was also found to contain a mixture of wild-type and different mutant alleles. Furthermore, the same mutations could be found within multiple strains (as defined by IS6110-based DNA fingerprinting) even when a single branch was examined (Fig. 2A). These observations support the hypothesis that INH-resistant SNPs arose multiple times by independent events in each phylogenetic branch and became common in the M. tuberculosis population through convergent evolution. A similar analysis was performed on INH-resistant SNPs in the inhA promoter and open reading frame (Fig. 2B) and in the aphC promoter (Fig. 2C). Again, we found that each INH-resistant-SNP allele was distributed on multiple phylogenetic branches, suggesting independent selection and convergent evolution. In total, 286 of 286 (100%) of the isolates containing INH-resistant SNPs (P = 0.0001 by the Fisher exact test) and 15 of the 15 (100%) different INH-resistant SNP alleles were distributed across the phylogenetic tree in patterns suggestive of multiple ancestry and consistent with convergent evolution (P = 0.001 by the Fisher exact test).
FIG. 2.
Distribution of INH-resistant SNPs. The location of each isolate containing the indicated drug resistance-associated SNPs was mapped to the phylogenetic tree shown in Fig. 1. The number of isolates harboring the mutation is shown for each allele (the number of DNA fingerprinting-defined strains is given in parentheses when it is less than the total number of isolates). (A) Mutations within katG315; (B) mutations within inhA (promoter and open reading frame); (C) mutations within the promoter region of ahpC (except for position −46, which is a non-drug-resistant SNP).
SNPs present in both INH-resistant and INH-susceptible isolates.
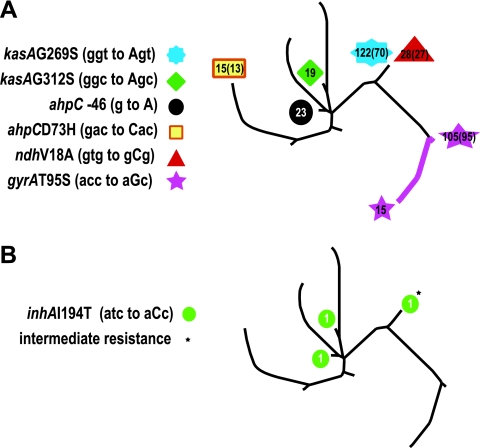
Some of the SNPs in our study had originally been thought to be responsible for INH resistance but were later found in both INH-resistant and INH-susceptible M. tuberculosis isolates, indicating that they did not have a role in resistance mechanisms (18). These “non-drug-resistance-associated SNPs” provided a good control group for our analysis of INH-resistant SNPs. Non-drug-resistance-associated SNPs included ahpC −46, ahpC(D73H), kasA(G269S), kasA(G312S), and ndh(V18A) mutations (see the supplemental material). As an additional control, we also analyzed each isolate for a mutation in codon 95 of gyrA, since this mutation is present within a fluoroquinolone antibiotic resistance-determining region but this mutation has been shown not to be related to antibiotic resistance (34).
When the distribution of the non-drug-resistance-associated SNPs was examined on the phylogenetic tree, we found that all 327 (100%) of the isolates containing non-drug-resistance-associated SNPs (P = 0.0001 by the Fisher exact test) and all 6 (100%) of the different non-drug-resistance-associated SNP alleles were monophyletic and thus showed no evidence of convergent evolution (Fig. 3A).
FIG. 3.
Distribution of non-drug-resistance-associated SNPs. The location of each isolate containing a SNP known to be found in both INH-resistant and INH-susceptible isolates is shown. The number of isolates harboring the mutation is shown for each allele (the number of DNA fingerprinting-defined strains is given in parentheses when it is less than the total number of isolates). (A) Mutations at positions 269 and 312 of kasA, −46 and 73 of ahpC, 18 of ndh, and 95 of gyrA. The colored line connecting the gyrA(T95S) isolates indicates that all isolates in these two SCGs contained this mutation, a result suggestive of a single mutational event in a common ancestor. (B) Mutations found in inhA at position 194 (I194T). The asterisk indicates the single isolate with an “intermediate” INH resistance phenotype; both of the other isolates were INH resistant.
Every non-drug-resistance-associated SNP was found within many different M. tuberculosis strains, indicating that the monophyletic distribution was not an epidemiological artifact caused by clonal disease outbreaks. The distribution of the non-drug-resistance-associated SNPs suggested that they each arose from a single common ancestor. This contrasted remarkably with the distribution of the INH resistance-associated SNPs (P = 0.0001 by the Fisher exact test). Our results provide strong statistical support for using convergent-evolution analysis as a test for significant mutations in putative drug-resistant genes.
A third type of SNP deserves special comment. Our study included three isolates with inhA(I194T) (atc-to-aCc) mutations. Two of these isolates were INH resistant, and one isolate was INH susceptible, suggesting that inhA(I194T) should be classified as a non-INH-resistance-associated mutation. However, the “INH-susceptible” inhA(I194T) isolate was noted to have significant growth on plates containing 0.1 μg/ml of INH, even though growth remained less than 1% of that of the no-antibiotic control. This culture became contaminated and could not be studied further. These results identify inhA(I194T) mutants as qualitatively different from the other non-INH-resistant isolates, and we have classified the single INH-susceptible isolate containing this mutation as having “intermediate” resistance (Fig. 3B; also data not shown). Interestingly, isolates containing this mutation had a phylogenetic distribution similar to that of the INH resistance-associated mutants shown in Fig. 2. We reanalyzed our results, this time including the inhA(I194T) mutation in the non-drug-resistance-associated SNP category, to exclude any possible biases caused by misassignment of this mutation. In this analysis, only 327 of (99%) of the 330 isolates containing non-drug-resistance-associated SNPs and 6 (83%) of the 7 non-drug-resistance-associated SNP alleles were monophyletic, with the difference in phylogenetic distribution between the INH resistance-associated SNPs and the non-drug-resistance-associated SNPs remaining highly significant (P = 0.001 by the Fisher exact test).
SNPs in the embB gene.
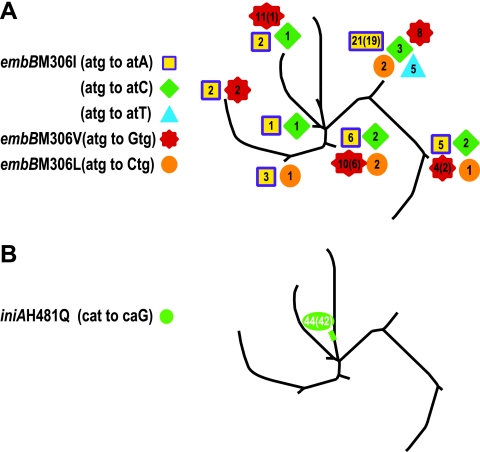
The embB gene is widely considered to be a target for the antituberculosis drug EMB. Mutations in embB306 were detected in many EMB-resistant M. tuberculosis isolates, and many groups consider embB306 mutations to be both a cause and a molecular marker for EMB resistance (26, 39). More recently, several groups have detected embB306 mutations in EMB-susceptible isolates, although these mutations were invariably found in association with resistance to at least one antituberculosis drug and usually in association with multidrug resistance (MDR) (17, 33). Thus, embB306 mutations had an unusual association with drug resistance, and the actual role of embB306 mutations in drug resistance was unclear. We tested each embB306 SNP allele for evidence of convergent evolution using the approach already described for INH-resistant SNPs (Fig. 4A). The results show that four of the five mutant embB306 alleles could be mapped to more than one phylogenetic branch. The embB(M306I) allele caused by an atg (Met) to atT (Ile) mutation represented the lone exception to this convergent-evolution pattern: all five isolates with this mutation were confined to SCG 5a. However, the two other embB alleles for this amino acid change, which involve a change of atg (Met) to atC or atA (Ile), as well as all other embB306 changes from the wild-type sequence, were widely distributed across the phylogenetic tree. Thus, it is possible that our failure to observe the atg-to-atT mutation in multiple SCGs was due to the small number of isolates carrying this mutation in our study sample. Together, these results provide strong evidence that most embB306 mutations provide a selective advantage to M. tuberculosis, likely through an association with drug resistance. Our findings provide population-based support for the allelic exchange studies recently performed by Safi et al. (31) with M. tuberculosis strain 210, demonstrating that embB306 mutations result in moderate increases in EMB MICs.
FIG. 4.
(A) Distribution of mutant embB306 alleles. The location of each isolate containing a mutant embB306 allele is shown. The number of isolates harboring the mutation is shown for each allele (the number of DNA fingerprinting-defined strains is given in parentheses when it is less than the total number of isolates). (B) Location of isolates containing a mutant iniA(H481Q) allele. The number of isolates harboring the mutation is shown, and the number of DNA fingerprinting-defined strains is given in parentheses.
Exploring the significance of SNPs in the iniBAC genes.
As seen in the case of embB306 mutations, the link between mutations and drug resistance phenotypes may not always be clear-cut. For example, some mutations may predispose to the development of drug resistance without, in themselves, conferring a fully drug-resistant phenotype. The genes of the M. tuberculosis iniBAC operon have been proposed to play such a role in both INH and EMB resistance. The wild-type iniA gene appears to be essential for the activity of an MDR-like pump that confers multidrug tolerance on M. tuberculosis while not affecting the MIC of any drug (8). Interestingly, a number of mutations in iniA, iniB, and iniC have also been described for INH- or EMB-resistant M. tuberculosis isolates, although each of these isolates also had another mutation that might explain the observed drug resistance phenotype (28, 29).
The prevalence of iniBAC mutations in our study sample permitted us to subject these mutations to phylogenetic analysis. We analyzed our entire collection of 1,003 isolates for the known mutations in iniBAC. Only the iniA(H481Q) mutation occurred at a sufficiently high frequency for phylogenetic analysis. The results showed that this mutation localized to a single branch (Fig. 4B). There were 44 isolates and 42 different DNA fingerprint patterns with this mutation, making it unlikely that this finding was due to chance or to the presence of a clonal outbreak. The results suggest that iniA(H481Q) mutations do not arise under selective pressure and are thus unlikely to contribute to drug resistance or to have a significant beneficial effect on the fitness of M. tuberculosis strains.
DISCUSSION
Our results and analyses of genes implicated in antibiotic resistance demonstrate the utility of phylogenetic analyses for identifying mutations likely to have a role in the biological fitness of M. tuberculosis. It is rather humbling to look a decade back to the early years of comparative genomics, when it was widely believed that the secret of pathogenicity could be quickly discovered by comparing the genomes of virulent and avirulent mycobacteria (for example, by comparing the virulent M. tuberculosis strain H37Rv to the avirulent strain H37Ra or by comparing M. tuberculosis to avirulent BCG). It has since become clear that comparative genomics only provides the starting point for pathogenicity studies. Genomic studies must be followed by allelic exchanges, gene knockouts, and often animal experiments to confirm which allelic variants have biological significance. In the case of M. tuberculosis, this involves a substantial investment in time and resources, because M. tuberculosis grows slowly and requires strict attention to biosafety. Furthermore, genetic manipulations are practically limited to a small number of laboratory or clinical strains, which may obscure important effects due to background variation in the clinical M. tuberculosis population. Our proposed phylogenetic approach circumvents these limitations by using simple SNP assays and by examining allelic variation in a large population of clinical M. tuberculosis isolates.
We demonstrated that SNPs in putative targets of INH resistance exhibit different phylogenetic distributions depending on whether these SNPs are strongly associated with INH resistance (and are likely to be the cause of INH resistance) or are not associated with INH resistance. INH resistance-associated SNPs appeared to have evolved multiple times in the M. tuberculosis populations, whereas the SNPs that were not associated with resistance were all monophyletic (i.e., limited to a single SCG) and appeared to have arisen only once. Our findings confirm the phylogenetic distribution of some katG and inhA promoter region mutations noted in two smaller studies (2, 4) and are consistent with the observation that the aphC −46 mutation can serve as a population genetic marker (5). However, our work is the first to systematically demonstrate the phylogenetic differences between INH resistance-associated and non-drug-resistance-associated SNPs and to propose a phylogenetic test for SNPs of uncertain function.
Our phylogenetic approach made it possible to test whether mutations in embB306 or iniA were likely to affect bacterial fitness. We detected strong convergent-evolution signals for virtually all embB306 mutations. These results support the hypothesis that embB306 mutations are linked to survival and growth in antibiotic environments despite the conflicting data from clinical laboratories. Thus, the current study may be used to justify the efforts required to perform definitive allelic exchange studies. In fact, allelic exchange studies performed in our laboratory do indeed suggest that some embB306 mutants have decreased susceptibility to EMB (31). Conversely, our results did not support the hypothesis that iniA(H481Q) mutations are worthy of further investigation. The large number of isolates and strains with this mutation available in our study sample made it highly unlikely that the observed monophyletic distribution of this mutation was due to chance alone. Rather, we conclude that this mutation shows no evidence of convergent evolution. The iniA(H481Q) mutation would normally have been a potentially interesting mutation, deemed worthy of molecular studies given its widespread distribution in clinical M. tuberculosis isolates, its previously reported association with INH and EMB resistance, and the known function of the iniA gene. Although it remains possible that the iniA(H481Q) mutation has an important biological function that is limited to the SCG 1 background, our phylogenetic approach strongly suggests that this SNP has a low priority with respect to studies of resistance.
Phylogenetic analysis is likely to become increasingly valuable as a tool for investigating the biological significance of SNPs as MDR and extensively drug resistant M. tuberculosis genomes are sequenced. Many of the mutations revealed by these sequencing studies have an uncertain role in the drug resistance phenotype (9, 13; Broad Institute, Mycobacterium tuberculosis Database [http://www.broad.mit.edu/annotation/genome/mycobacterium_tuberculosis_spp/MultiHome.html]), and a phylogenetic approach may help identify a subset of mutations that are particularly worthy of additional study. Furthermore, our analytic approach does not necessarily have to be limited to studies of antibiotic resistance. Antibiotic resistance served as a good model for this investigation because the resistance phenotype was relatively easy to confirm and the order of SNP acquisition (i.e., from “wild type” to “mutant”) could be inferred from the historical record showing antibiotic resistance as a recent phenomenon. However, it is likely that all mutations that provide strong selective advantages to clinical M. tuberculosis isolates will exhibit similar behavior. Thus, our approach may be used to screen entire classes of SNPs for biological relevance on a much wider scale. Finally, the phylogenetic approach for fitness mutations can be widely applied to studies of organisms in addition to M. tuberculosis. This type of analysis will be of little use for bacteria with high rates of lateral gene exchange or high mutation rates, such as Helicobacter pylori (12). However, a number of clinically important or scientifically interesting organisms, such as Bacillus anthracis or Francisella tularensis (7, 30), that have highly conserved genomes should be amenable to our approach.
Supplementary Material
Acknowledgments
This work was supported by grants R01AI46669 and R01AI49352. T.S.W. was supported in part by NIH research contract N01-AI-30058 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print on 30 June 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Alland, D., D. W. Lacher, M. H. Hazbon, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45:39-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbón, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azhikina, T., N. Gvozdevsky, A. Botvinnik, A. Fushan, I. Shemyakin, V. Stepanshina, M. Lipin, C. Barry III, and E. Sverdlov. 2006. A genome-wide sequence-independent comparative analysis of insertion-deletion polymorphisms in multiple Mycobacterium tuberculosis strains. Res. Microbiol. 157:282-290. [DOI] [PubMed] [Google Scholar]
- 4.Baker, L., T. Brown, M. C. Maiden, and F. Drobniewski. 2004. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg. Infect. Dis. 10:1568-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, L. V., T. J. Brown, O. Maxwell, A. L. Gibson, Z. Fang, M. D. Yates, and F. A. Drobniewski. 2005. Molecular analysis of isoniazid-resistant Mycobacterium tuberculosis isolates from England and Wales reveals the phylogenetic significance of the ahpC −46A polymorphism. Antimicrob. Agents Chemother. 49:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brimacombe, M., M. Hazbon, A. S. Motiwala, and D. Alland. 2007. Antibiotic resistance and single-nucleotide polymorphism cluster grouping type in a multinational sample of resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 51:4157-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broekhuijsen, M., P. Larsson, A. Johansson, M. Bystrom, U. Eriksson, E. Larsson, R. G. Prior, A. Sjostedt, R. W. Titball, and M. Forsman. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colangeli, R., D. Helb, S. Sridharan, J. Sun, M. Varma-Basil, M. H. Hazbon, R. Harbacheuski, N. J. Megjugorac, W. R. Jacobs, Jr., A. Holzenburg, J. C. Sacchettini, and D. Alland. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55:1829-1840. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.de Jong, B. C., P. C. Hill, R. H. Brookes, S. Gagneux, D. J. Jeffries, J. K. Otu, S. A. Donkor, A. Fox, K. P. McAdam, P. M. Small, and R. A. Adegbola. 2006. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J. Infect. Dis. 193:1279-1286. [DOI] [PubMed] [Google Scholar]
- 11.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbón, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald, J. R., and J. M. Musser. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9:547-553. [DOI] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutacker, M. M., B. Mathema, H. Soini, E. Shashkina, B. N. Kreiswirth, E. A. Graviss, and J. M. Musser. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193:121-128. [DOI] [PubMed] [Google Scholar]
- 15.Hanekom, M., G. D. van der Spuy, E. Streicher, S. L. Ndabambi, C. R. McEvoy, M. Kidd, N. Beyers, T. C. Victor, P. D. van Helden, and R. M. Warren. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 45:1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazbón, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazbón, M. H., M. Bobadilla del Valle, M. I. Guerrero, M. Varma-Basil, I. Filliol, M. Cavatore, R. Colangeli, H. Safi, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. Garcia-Garcia, A. Davidow, M. Brimacombe, C. I. Leon, T. Porras, M. Bose, F. Chaves, K. D. Eisenach, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, and D. Alland. 2005. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob. Agents Chemother. 49:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazbón, M. H., M. Brimacombe, M. Bobadilla Del Valle, M. Cavatore, M. I. Guerrero, M. Varma-Basil, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. Garcia-Garcia, C. I. Leon, M. Bose, F. Chaves, M. Murray, K. D. Eisenach, J. Sifuentes-Osornio, M. D. Cave, A. Ponce de Leon, and D. Alland. 2006. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 50:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen, I. S., V. O. Thomsen, M. Marjamaki, A. Sosnovskaja, and B. Lundgren. 2004. Rapid, automated, nonradiometric susceptibility testing of Mycobacterium tuberculosis complex to four first-line antituberculous drugs used in standard short-course chemotherapy. Diagn. Microbiol. Infect. Dis. 50:103-107. [DOI] [PubMed] [Google Scholar]
- 20.Kapetanaki, S. M., S. Chouchane, S. Yu, X. Zhao, R. S. Magliozzo, and J. P. Schelvis. 2005. Mycobacterium tuberculosis KatG(S315T) catalase-peroxidase retains all active site properties for proper catalytic function. Biochemistry 44:243-252. [DOI] [PubMed] [Google Scholar]
- 21.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, Atlanta, GA.
- 22.Kong, Y., M. D. Cave, L. Zhang, B. Foxman, C. F. Marrs, J. H. Bates, and Z. H. Yang. 2007. Association between Mycobacterium tuberculosis Beijing/W lineage strain infection and extrathoracic tuberculosis: insights from epidemiologic and clinical characterization of the three principal genetic groups of M. tuberculosis clinical isolates. J. Clin. Microbiol. 45:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, C. N., and L. B. Heifets. 1987. Determination of minimal inhibitory concentrations of antituberculosis drugs by radiometric and conventional methods. Am. Rev. Respir. Dis. 136:349-352. [DOI] [PubMed] [Google Scholar]
- 24.Manca, C., L. Tsenova, S. Freeman, A. K. Barczak, M. Tovey, P. J. Murray, C. Barry, and G. Kaplan. 2005. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J. Interferon Cytokine Res. 25:694-701. [DOI] [PubMed] [Google Scholar]
- 25.Mendelson, M., S. Walters, I. Smith, and G. Kaplan. 2005. Strain-specific mycobacterial lipids and the stimulation of protective immunity to tuberculosis. Tuberculosis (Edinburgh) 85:407-413. [DOI] [PubMed] [Google Scholar]
- 26.Plinke, C., S. Rusch-Gerdes, and S. Niemann. 2006. Significance of mutations in embB codon 306 for prediction of ethambutol resistance in clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 50:1900-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramaswamy, S. V., A. G. Amin, S. Goksel, C. E. Stager, S. J. Dou, H. El Sahly, S. L. Moghazeh, B. N. Kreiswirth, and J. M. Musser. 2000. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:326-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramaswamy, S. V., R. Reich, S. J. Dou, L. Jasperse, X. Pan, A. Wanger, T. Quitugua, and E. A. Graviss. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 31.Safi, H., B. Sayers, M. H. Hazbon, and D. Alland. 2008. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob. Agents Chemother. 52:2027-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 33.Shen, X., G. M. Shen, J. Wu, X. H. Gui, X. Li, J. Mei, K. Deriemer, and Q. Gao. 2007. The association between embB codon 306 mutations and drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 51:2618-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telenti, A., N. Honore, C. Bernasconi, J. March, A. Ortega, B. Heym, H. E. Takiff, and S. T. Cole. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theus, S. A., M. D. Cave, K. Eisenach, J. Walrath, H. Lee, W. Mackay, C. Whalen, and R. F. Silver. 2006. Differences in the growth of paired Ugandan isolates of Mycobacterium tuberculosis within human mononuclear phagocytes correlate with epidemiological evidence of strain virulence. Infect. Immun. 74:6865-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tracevska, T., I. Jansone, A. Nodieva, O. Marga, G. Skenders, and V. Baumanis. 2004. Characterisation of rpsL, rrs and embB mutations associated with streptomycin and ethambutol resistance in Mycobacterium tuberculosis. Res. Microbiol. 155:830-834. [DOI] [PubMed] [Google Scholar]
- 38.van Doorn, H. R., E. C. Claas, K. E. Templeton, A. G. van der Zanden, A. te Koppele Vije, M. D. de Jong, J. Dankert, and E. J. Kuijper. 2003. Detection of a point mutation associated with high-level isoniazid resistance in Mycobacterium tuberculosis by using real-time PCR technology with 3′-minor groove binder-DNA probes. J. Clin. Microbiol. 41:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilcheze, C., F. Wang, M. Arai, M. H. Hazbon, R. Colangeli, L. Kremer, T. R. Weisbrod, D. Alland, J. C. Sacchettini, and W. R. Jacobs, Jr. 2006. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat. Med. 12:1027-1029. [DOI] [PubMed] [Google Scholar]
- 41.Wei, C. J., B. Lei, J. M. Musser, and S. C. Tu. 2003. Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob. Agents Chemother. 47:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.