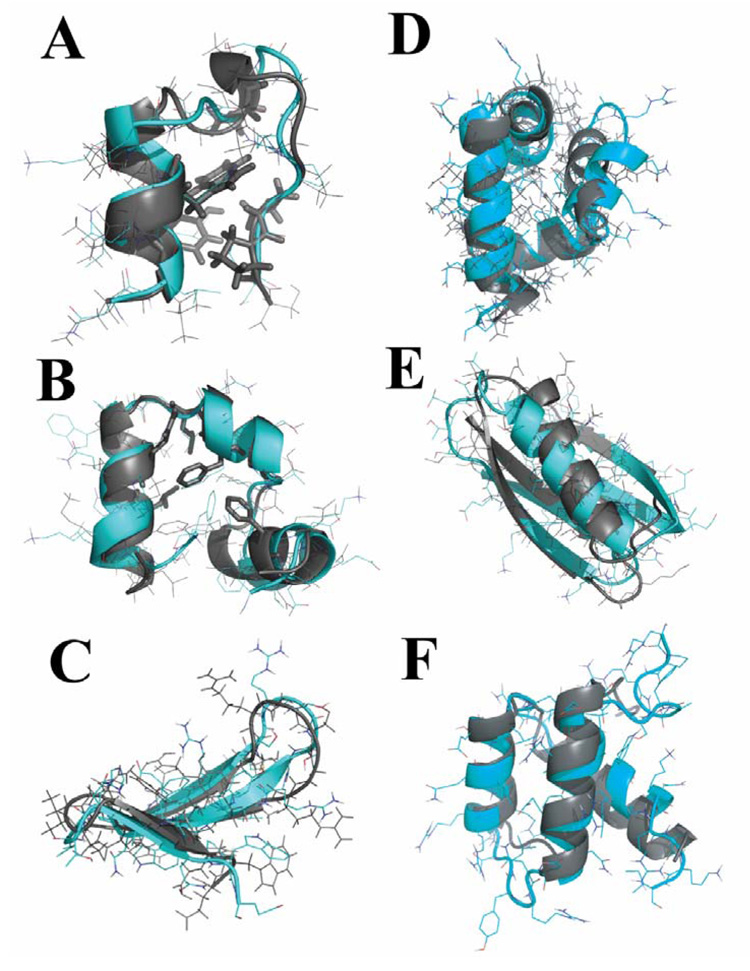
Figure 1. Ab initio folding of six small proteins in all-atom DMD simulations.
(A) In the lowest-RMSD Trp-cage structure from simulations, the protein core is well packed and the sidechain rotamers of core residues (in stick representation) are also consistent with the NMR structure. The structure from simulations is in cyan and the one determined by experiments is colored in gray. The same color code is used in the following panels. (B) In simulations of villin headpiece, we find that the protein consistently folds to its native state with an average RMSD of 2–3Å. The core residues Phe6, Phe17, Leu20, Gln25, and Leu28 are as closely packed against each other as they are observed in the crystal structure. For the WW domain (C) and engrailed homeodomain (D), the secondary structures from simulations align well with respect to the experimentally determined structures except that loops have larger deviations. In the simulations of GB1 (E) and bacterial ribosomal protein L20 (F), the near native states are observed in DMD simulations.