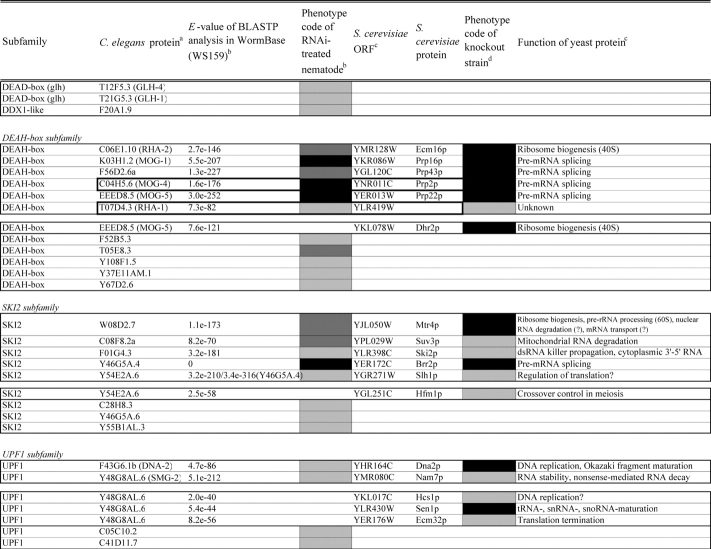
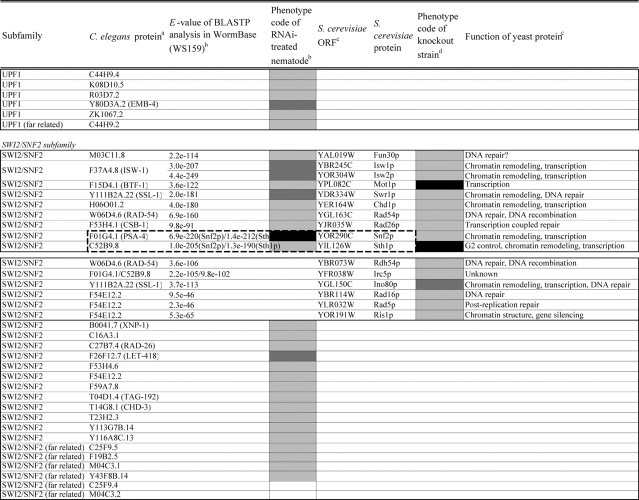
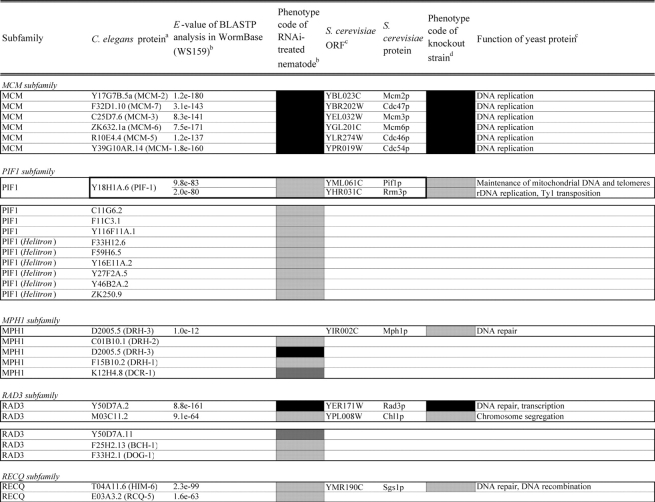
Table 2.
Comparison of loss-of-function phenotypes of helicase-like genes in S. cerevisiae and C. elegans
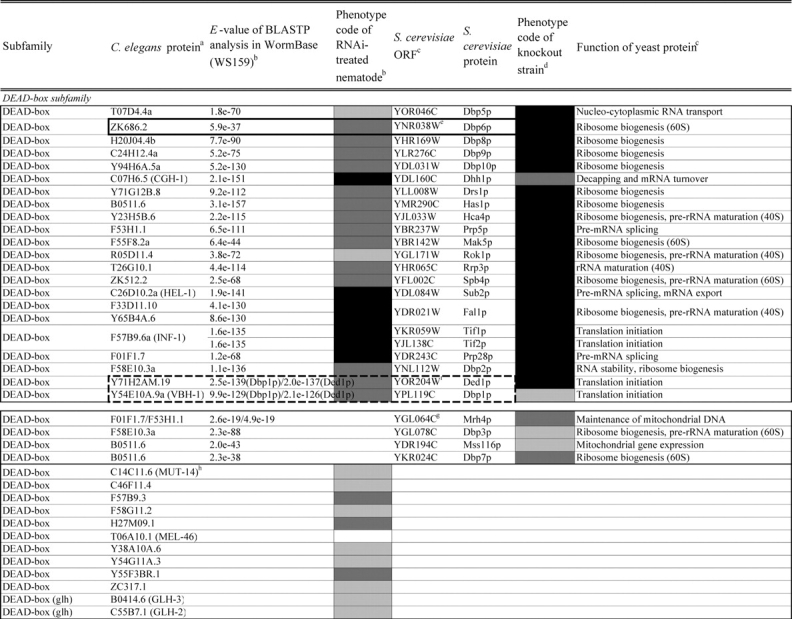
Loss of function phenotypes of helicase like proteins in S. cerevisiae and C. elegans are summarized according to subfamily for comparison between species. aC. elegans proteins putatively orthologous to the yeast helicase like proteins and E values of the BLAST analyses are shown. C. elegans proteins were identified by BLASTP analysis and database search against the InParanoid database (version 4.0 updated April 2005, http://inparanoid.cgb.ki.se/). Several putative orthologs were identified as reciprocal best BLAST hits with an E val ue <1.0e 30 between S. cerevisiae and C. elegans. Suffixes ‘a’ and ‘b’ indicate variants with the highest homology to the yeast protein. bPhenotype code (C. elegans): The phenotypes of RNAi treated nematodes are indicated by gray scale coding: Emb in black, Lva and Gro in dark gray, and WT (no phenotype) in light gray. Empty code: no data (not tested). A phenotype code for the most intense phenotype is indicated. cClassification and functions of yeast helicase like proteins are according to the yeast RNA helicase database by Linder and colleagues. dPhenotypes of the corresponding knockout strains were mainly obtained from the Sacch aromyces Genome Database and our previous report8 and shown by phenotype codes: lethal in black, slow growth in dark gray, and viable in light gray, no data in white. eThe proteins surrounded with bold lines are a putative orthologous pair based on BLASTP scores, but were not in the InParanoid database. The Ku70 and Ku80 homologs in yeast and nematodes are described in the Saccharomyces Genome Database and WormBase. fTwo pairs of yeast proteins (Snf2p and Sth1p, Ded1p and Dbp1p) with two C. elegans orthologs are surrounded by dashed lines. gThe yeast proteins with BLAST scores lower than that of the putative homologs or without any sequence homologies to C. elegans proteins are indicated in separated box for each subfamily. hC. elegans proteins without significant similarities to yeast helicase like proteins are also indicated separately. iND, not detected . Several C. elegans proteins with E values greater than 1e 10 when compared with the Y’ Hel1 proteins were omitted because the similarities were to low complexit y regions in the amino acid sequences. jTwenty five budding yeast specific proteins including subtelomere specific helicase like proteins and four yeast proteins (Hrq1p, Hpr5p, Hmi1p, and Irc3p) and six C. elegans (higher eukaryot e) specific proteins (DIC 1, NSH 1, POLQ 1, F46G11.1, F52G3.3, and F52G3.4) were detected.