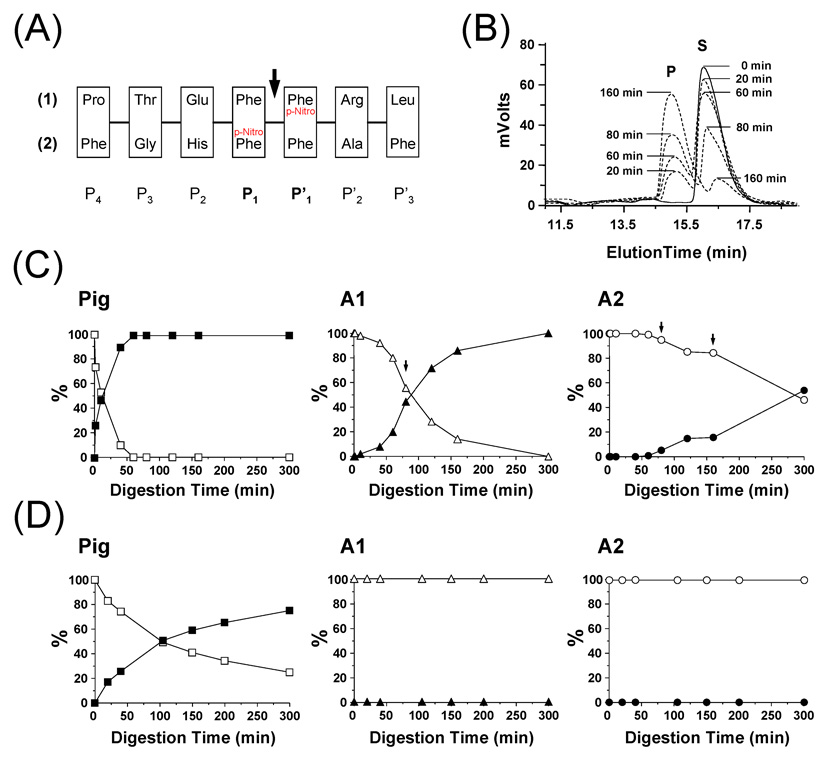
Figure 4.
Analysis of pepsin digestion of synthetic test peptides. (A) Amino-acid sequence of the two synthetic peptides used in this study. The P4 to P’3 nomenclature refers to the position of each residue. The arrow indicates the position of the scissile peptide bond between P1 and P’1. (B) Example of the HPLC elution profile observed at 280 nm for A1 digestion of synthetic peptide #1 (Legend: S = Substrate; P = Product). The digestion was performed in 100 mM sodium acetate buffer, pH 2.5, 0 °C using a 1 to 10 protease/substrate ratio (w/w). At defined time points, 600 pmoles were loaded onto a Jupiter 4u Proteo 90A column and peptides were eluted using a 5 to 70 % ACN gradient in 20 min. (C) Digestion of synthetic peptide #1 by pig pepsin, A1 and A2. Solid and empty symbols correspond to the substrate and the product, respectively. Arrows indicate time points at which additional amounts of fish pepsins were added [60 min: 4 µg (A1 and A2); 160 min: 4 µg (A2)]. (D) Digestion of synthetic peptide #2 by pig pepsin, A1 and A2. No digestion was observed with A1 and A2. Solid and empty symbols correspond to the substrate and the product, respectively.