Abstract
Cytochrome P450s exist ubiquitously in all organisms and are involved in many biological processes. Allene oxide synthase (AOS) is a P450 enzyme that plays a key role in the biosynthesis of oxylipin jasmonates, which are involved in signal and defense reactions in higher plants. The crystal structures of guayule (Parthenium argentatum) AOS (CYP74A2) and its complex with the substrate analog 13(S)-hydroxyoctadeca-9Z,11E-dienoic acid have been determined. The structures exhibit a classic P450 fold but possess a heme-binding mode with an unusually long heme binding loop and a unique I-helix. The structures also reveal two channels through which substrate and product may access and leave the active site. The entrances are defined by a loop between β3–2 and β3–3. Asn-276 in the substrate binding site may interact with the substrate's hydroperoxy group and play an important role in catalysis, and Lys-282 at the entrance may control substrate access and binding. These studies provide both structural insights into AOS and related P450s and a structural basis to understand the distinct reaction mechanism.
Keywords: oxylipin, jasmonate, guayule
Cytochrome P450s are one of the largest superfamilies of enzymes and are ubiquitously distributed in all biological organisms. They are heme-containing enzymes catalyzing a wide range of chemical reactions (1, 2). P450s play very important roles in drug metabolism and detoxification for humans and animals and are key players in the synthesis of natural products, such as antibiotics in microorganisms and a broad range of secondary metabolites in plants. Allene oxide synthase (AOS) is a P450 enzyme that plays a key role in the biosynthesis of oxylipins, bioactive compounds involved in signal and defense reactions in higher plants, mammals, and algae (3, 4). Jasmonates are plant oxylipins and ubiquitous plant growth regulators and are synthesized via the AOS branch of the lipoxygenase (LOX) pathway. AOS catalyzes a dehydration reaction to convert 13(S)-hydroperoxide (Fig. 1), derived from linolenic acid by LOX, to allene oxide, which is further cyclized by allene oxide cyclase, leading to the formation of jasmonic acid. Structures of Arabidopsis thaliana allene oxide cyclase and some LOXs have been reported (5, 6), but no AOS structure has been characterized to date.
Fig. 1.
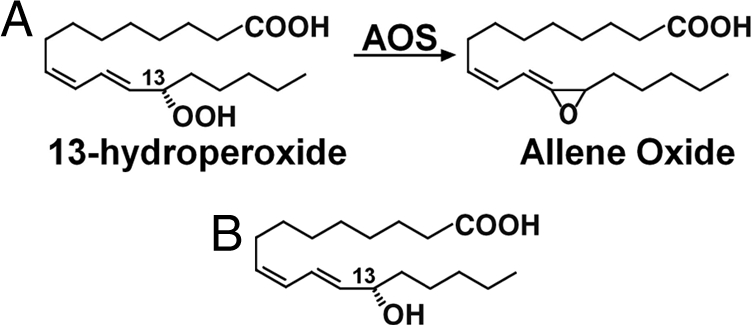
Reaction catalyzed by AOS. (A) With substrate 13(S)-HPOD. (B) Structure of substrate analog 13(S)-HODE.
Guayule (Parthenium argentatum) can accumulate an extraordinarily large number of rubber particles. AOS from guayule is the most abundant protein associated with rubber particles. It compromises ≈50% of the proteins extracted from guayule rubber particles, which was found to be a P450 enzyme with AOS activity (7), similar to the AOS identified from flaxseed (8, 9). However, the possible involvement of such an active enzyme in rubber biosynthesis remains unclear.
Although most AOSs from plants are membrane-associated, AOSs from guayule and corn, and recombinant AOSs from barley and tomato, have no N-terminal membrane anchor and are cytosolic (9, 10). A distinctive feature of guayule AOS is the absence of the putative transit sequence, which is present in flaxseed AOS and others for targeting to plastids or mitochondria.
AOSs are peroxide-metabolizing P450s and belong to the CYP74A group. They are atypical P450s, are self-sufficient, and do not require molecular oxygen and NADPH reductase. AOSs use an acyl hydroperoxide both as the substrate and the oxygen donor. The typical class I and II P450s require electron transfer partners, i.e., FAD-containing reductase and an iron sulfur redoxin (for class I), and a P450 reductase (for class II). There is another class of P450s that receive electrons directly from NAD(P)H (2). The biochemical mechanisms underlying P450 reactions, especially in biosynthesis, are not well understood.
AOS from guayule has been classified as CYP74A2 with enzymatic activity toward 13(S)-hydroperoxide (7) but not 9(S)-hydroperoxide (Z.P. unpublished results). All CYP74A subfamily enzymes use 13(S)-hydroperoxides as substrates and are named 13-AOS (11). There are two other types of AOSs with different substrate specificities. The 9/13-AOS enzymes from barley and rice recognize both 9(S)-hydroperoxides and 13(S)-hydroperoxides (12). The 9-AOSs from tomato and potato have specificity for 9(S)-hydroperoxides (13, 14). The 9/13- and 9-AOS are grouped in the CYP74C subfamily.
Structural studies of P450s provide an essential basis for understanding their complex catalytic reactions. To date, many structures have been reported for soluble bacterial/mitochondrial class I P450s (15–21). Class II enzymes in eukaryotes are microsomal P450s containing a membrane-binding anchor at the N terminus. Several mammalian microsomal P450 structures have been solved, including the closely related rabbit P450s 2C5 and 2B4, and the human P450s 2C8, 2C9, 2A6, 3A4, and 2D6 (22–29). Structures have also been reported for other classes P450s, e.g., the fungal nitric oxide reductase P450nor (30), and prostacyclin synthases (PGIS, CYP8A1), which are involved in endoperoxide metabolism to catalyze the isomerization of prostaglandin H2 to prostacyclin (31, 32). A very large number of P450s are present in plants, with nearly 2,900 named plant P450s in the P450 database (http://drnelson.utmem.edu/CytochromeP450.html). Many are known to be involved in the biosynthesis of natural products. So far, no plant P450 structure has been reported.
Based on amino acid sequence information, AOS contains an unusual heme-binding region and possesses a defective I-helix without the highly conserved threonine, suggesting an altered I-helix function (7). Homology modeling of AOSs is difficult because of the very poor sequence identity with any other P450 enzymes. Detailed structural information is therefore essential to understand the catalytic mechanism of AOS. Recently we successfully expressed, purified, and crystallized AOS (33).
Here, we report crystal structures of AOS (CYP74A2) from guayule (P. argentatum) and its complex with the substrate analog 13(S)-hydroxyoctadeca-9Z,11E-dienoic acid [13(S)-HODE]. These plant P450 crystal structures revealed distinct structural features of AOS and its interactions with heme and substrate analog. From the structures, a heme-binding mode and two access channels for substrate to enter and/or for product to leave the active site have been identified, along with residues important for substrate binding and catalysis.
Results and Discussion
Structure Determination and Overall Structure.
The crystal structures of AOS in native form (Native 1) and in complex with the substrate analog 13(S)-HODE were determined by using the multiple isomorphous replacement with anomalous scattering (MIRAS) method. Another AOS native crystal form with a small cell unit, denoted as Native 2, diffracted to 1.8-Å resolution, and the structure was determined by using the molecular replacement (MR) method [supporting information (SI) Tables S1 and S2].
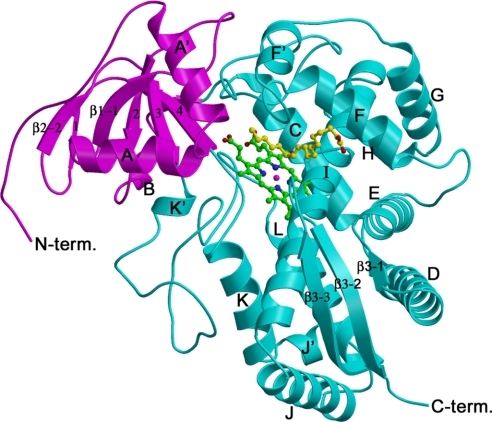
The structure of AOS contains two domains with three β-sheets and 15 α-helices (Fig. 2). The major α-domain is predominantly α-helices and contains a conserved structural core around heme, whereas the small β-domain is predominantly β-sheets with two α-helices A′ and A in the N terminus, and a 310 helix B.
Fig. 2.
Ribbon diagram of the structure of AOS with bound heme and 13(S)-HODE. The α- and β-domains are shown in cyan and magenta with the secondary structures and the N and C termini labeled. The heme and 13(S)-HODE molecules are shown as ball-and-stick models. Figs. 2, 3, 4A, and 5 were prepared with MOLSCRIPT (45) and RASTER3D (46).
AOS is structurally similar to other P450s despite the very low sequence identity (Figs. S1 and S2). Superimposing the structures of AOS and human PGIS [Protein Data Bank (PDB) ID code 2IAG] by using the DaliLite program (34) gave a rmsd of 3.9 Å with 401 residues aligned and 14% sequence identity. Superimposing the structures of AOS and human P450 2C9 (class II; PDB ID code 1OG5) gave a rmsd of 3.6 Å with 394 residues aligned and 12% sequence identity. Interestingly, superimposing the structures of AOS and bacterial class I P450BM-3 (PDB ID code 2HPD) gave a lower rmsd (3.0 Å) with 408 residues aligned and 13% sequence identity. The differences between AOS and PGIS and other P450s are observed widely (Fig. 3 and Figs. S1 and S2), and the distinct structural features of AOS are mainly related to its unique heme-binding mode, substrate recognition, and specificity (see below).
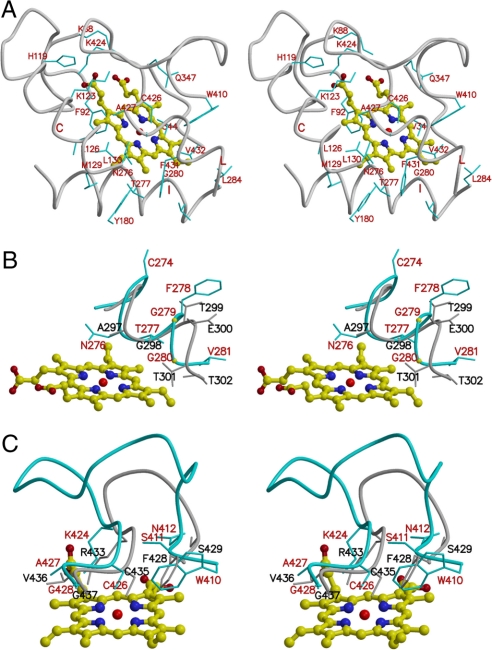
Fig. 3.
Heme-binding site. (A) Stereo diagram shows heme molecule and its interactions with AOS. The structure of heme is shown as a ball-and-stick model. (B) Stereo diagram shows a comparison of I-helix of AOS (in cyan) and human P450 2C9 (in gray). (C) Stereo diagram shows a comparison of heme-binding loop of AOS (in cyan) and human P450 2C9 (in gray).
The native AOS structures in the two different crystal forms were nearly identical with a rmsd of 0.33 Å with 466 Cα atoms aligned. The major differences observed include a loop region between β3–2 and β3–3, and the five N-terminal residues that are well defined in the small-cell crystal form (i.e., Native 2), but disordered in the crystal form 1 with a large-unit cell. The AOS molecules are packed more tightly and much better ordered in the small-cell crystal form with a much lower average temperature factor (20.9 Å2), whereas the average value was 42.9 Å2 for the AOS molecule in the large-cell crystal form.
The structure of AOS bound with substrate analog is also nearly identical to that of native AOS, with a rmsd of 0.22 Å over all Cα atoms. This difference is even smaller than that between structures in different crystal forms, indicating that substrate does not induce a significant conformational change in AOS.
Binding Mode with Heme.
Unique I-helix.
The heme prosthetic group is located mainly between helices I and L in the AOS structure (Figs. 2 and 3). Cysteine-426 acts as the fifth ligand for the iron of the heme cofactor. The classic P450 monooxygenases contain a signature sequence (A/G-G-X-D/E-T-T/S) in helix I, regarded as an oxygen-binding motif with the conserved glycine pointing at the center of the heme and the conserved threonine pointing to the oxygen-binding site. AOS does not require molecular oxygen and contains a unique I-helix different from other P450s (Fig. 3B), with a distinctive sequence (A276-T277-X-G279-G280-X) in its central region. A threonine and glycine (Thr-277 and Gly-280 in AOS) are present at the positions corresponding to the conserved glycine and threonine of bacterial and mammalian P450s, and an asparagine (i.e., Asp-276 in AOS) is present at the corresponding position of the conserved alanine/glycine (e.g., Ala-297 in human P450 2C9; Fig. 3B). In the AOS structure, the residue Gly-280 is ≈4 Å from the vinyl group of the heme pyrrole ring A. Thr-277 is <3.5 Å from the methyl and vinyl groups of heme pyrrole ring B, and the side chain of Asn-276 is <4 Å from heme pyrrole ring C. The threonine, glycine, and asparagine residues are highly conserved in AOSs and the CYP74 family (Fig. S3). The adjacent residues Phe-275 and Lys-282 in the N and C termini are also highly conserved. This sequence (F-N-T/S-X-G-G-X-K) may be regarded as a signature for this family and plays very important roles in heme binding and catalytic function.
Compared with other classes of P450s, the helix I in AOS has an extra amino acid (Cys274 in our alignment, Fig. S1) in the middle, and the central part of the helix is distorted with a disturbed hydrogen bond pattern. With this extra amino acid, the adjacent residues may be further extended to the active site.
Unusual heme-binding loop.
The heme-binding loop is a distinct structural feature of AOS related to its unique heme-binding mode. It is located on the proximal face of the heme. In other P450s, this loop usually forms a compact β-turn, is located over the heme in the proximal side, and contains the most typical P450 consensus sequence, F-X-X-G-X-R-X-C-X-G. Compared with human P450 2C9, PGIS, and other P450s (Fig. 3C and Fig. S1), the corresponding loop (S411–G428) in AOS is unusually long with eight more amino acids. The C-terminal portion of the heme-binding loop in AOS is conserved with a similar basic residue (lysine) and two identical residues cysteine and glycine (i.e., K-X-C-X-G) that are present in the similar positions of the corresponding residues in human P450 2C9. Besides Cys-426 interacting with the heme iron as the fifth ligand, Ala-427 and Gly-428 form hydrophobic interactions with the heme and are <4 Å from the heme pyrrole rings C and B, respectively. Lys-424 forms a strong charge–charge interaction with the propionate group of the heme pyrrole ring C.
The major difference between AOS and other P450s is the middle portion of the heme-binding loop (Fig. 3C and Fig. S1), which is extended to the surface with eight more amino acids in AOS, interacts with other parts of the enzyme and covers the internal portion of the loop that interacts with the heme. In the N-terminal portion of the heme-binding loop, although the sequence is not conserved, the first two N-terminal amino acids are located in similar positions. In the AOS structure, a Ser-411 is located in the position corresponding to the first phenylalanine in the typical P450 consensus sequence. Interestingly, the indole ring of the adjacent residue, Trp-410 in AOS, is present in the position occupied by the benzyl side chain of the phenylalanine, providing a similar hydrophobic support for heme.
The region between “meander” and the heme-binding loop is also different between AOS and other P450s. Following the conserved meander, AOS contains a 310 helix (i.e., G399–Y407), whereas PGIS has a small β-sheet.
There are other unique interactions between enzyme and heme in the AOS structure. The propionate group of the heme pyrrole ring C also forms charge–charge interactions with His-119 and Lys-123 in helix C. The propionate group of pyrrole ring D interacts with Lys-88. Including the Lys-424 mentioned above, there are four basic residues forming strong charge–charge interactions with heme. These strong and extensive interactions may make the heme group very stable and more restricted in the active site. For other P450s, there are usually only one or two charge–charge interactions formed between enzyme and heme.
All of these structural features define a heme-binding mode for AOS, which is quite different from that of PGIS and other classes of P450s.
Substrate-Binding Site and Access Channels.
The substrate analog 13(S)-HODE is very similar to the substrate 13(S)-hydroperoxy-9(Z),11(E)-octadecadienoic acid [13(S)-HPOD], with a hydroxyl group in the C13 position, in place of the hydroperoxyl group of the substrate (Fig. 1). In the structure of AOS, the 13(S)-HODE molecule was located in a very narrow and deep pocket observed on the distal side of the heme (Fig. 4A and B and Fig. S4). This substrate-binding pocket is formed by helices F and I, and loops between helix C and β1–5, between helices F and F′, between helix K and β1–4, and between β3–2 and β3–3, and consists mainly of hydrophobic residues such as Phe-92, Phe-275, and Phe-278. Interestingly, a polar residue (Asn-276) is present in the active site and is very close to heme. This asparagine residue is conserved in all AOSs (Fig. 3) but not in other P450s (Fig. 3B and Fig. S1), suggesting a key role specific for AOS catalysis and function.
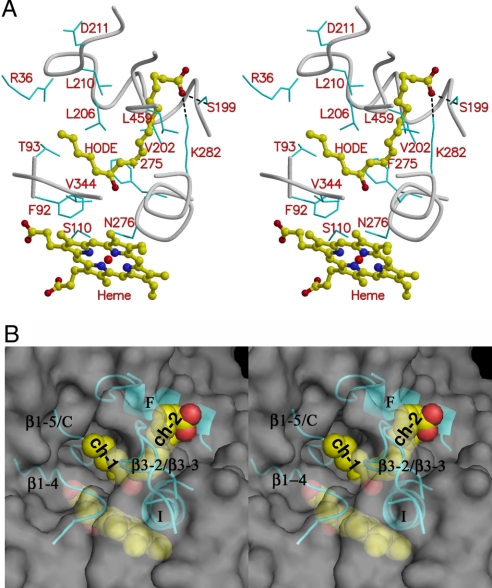
Fig. 4.
Substrate-binding pocket. (A) Stereo diagram shows 13(S)-HODE and its interactions with AOS. 13(S)-HODE and heme are shown as ball-and-stick models. Some protein residues in the substrate-binding pocket are labeled and shown in cyan as bond models. (B) Stereo diagram of semitransparent molecular surface of AOS showing substrate-binding site and the two access channels labeled as ch.1 and ch.2. Both 13(S)-HODE (Upper) and heme (Lower) are shown as sphere models. The helices F and I, strand β1–4, and some loops (i.e., loops between β1–5 and helix C, between helix K and β1–4, and between β3–2 and β3–3) around the substrate analog are shown as ribbons. This molecular surface figure was produced with PyMOL (http://pymol.sourceforge.net/).
The loop F/F′ and the loop between β3-2 and β3-3 are very close to each other on the surface of the substrate-binding pocket, and they define two entrances and channels in both sides, connecting the active site to the surface (Fig. 4A and B). One channel, designated as channel 1, is mainly between helices F and I. Another channel, channel 2, is between helix F and strand β1-4 with helix A in the entrance. The long fatty acid chain of 13(S)-HODE bends at the C13 position with the 13-hydroxyl group pointing to the heme iron; the long α-chain with the carboxyl group is in channel 1, and the short ω-chain with the methyl group is in channel 2.
Access channels have been observed in other P450 structures in quite different locations and formations. For P450cam, there is one long deep access channel in the open form structure (PDB ID 1K2O) (35), and the channel may be covered in the closed form (e.g., 1DZ4). In zebrafish PGIS, two access channels were observed (32) but in totally different locations; channel 1 is in a near opposite direction with its entrance near helix B′ and the loop B′/C on the other side of the surface, compared with channel 1 in AOS. Channel 2 is also different with an entrance near the helix A′ and β-sheet 1, whereas the entrance of access channel 2 in AOS is near the loop A′/A and the loop between β3–2 and β3–3. Thus, AOS possesses two special access channels quite different from those in other P450s.
Substrate Recognition.
Six putative substrate recognition sites (SRS1–6) for P450s have been proposed based on the analysis of the CYP2 family and P450cam structure (36). The substrate-binding pocket in AOS may also be classified into similar substrate recognition sites although it is quite different from other P450s in sequence and structure.
The loop region before helix C was identified as the SRS1 site in other P450s. In the AOS structure, the corresponding region is a very long loop between strand β1–5 and helix C with 10 more amino acids than in PGIS. The N-terminal portion near Thr-95 is closer to the β-sheet 1, and the regions near Phe-92 and Ser-110 are closer to heme and extend further into the substrate binding pocket, whereas PGIS contains a short helix in the corresponding regions that is further away from the heme. In the AOS–substrate analog structure, residues Phe-92 and Thr-93 are close to the short ω-chain of 13(S)-HODE (Fig. 4A). Ser-110 forms a hydrogen bond with Asn-276, which may interact with the hydroperoxyl group of the substrate (see below, Fig. 5).
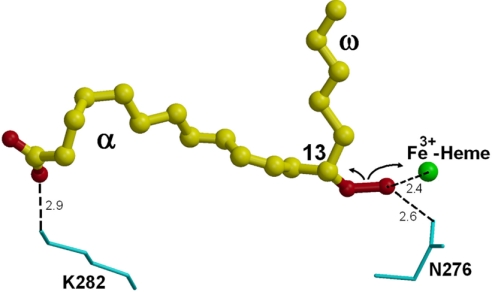
Fig. 5.
Putative substrate-binding mode and the heme-assisted homolytic scission. The substrate 13(S)-HPOD is shown as ball-and-stick models, and its oxygen atoms (i.e., the hydroperoxy and carboxyl groups) are shown in red. Residues Asn-276 and Lys-282 are labeled and shown in cyan as bond models. Heme iron is shown as a sphere model in green. The possible interactions are indicated by the dashed lines and approximate distances (Å).
In other P450s, the C-terminal region of helix F and the N-terminal region of helix G were identified as SRS2 and SRS3, respectively, and the loop F/G is present as a short β-turn to link the helices. In AOS, the helix F is on the molecular surface and at the entrance of the substrate-binding pocket, and the link between helices F and G is much longer with an extra helix F′ and 12 more residues than in PGIS, and the loop F/F′ turns nearly 90° toward helix A′ and β-sheet 3. The helix F and the following loop F/F′ form parts of both the two access channels, and extensively interact with both α- and ω-chains of 13(S)-HODE. Besides the major hydrophobic interactions, Ser-199 in helix F forms a hydrogen bond with the carboxyl group of the substrate analog (Fig. 4). Thus, as in other P450s, helix F is important for interacting with substrate as a part of the SRS2. The following loop and helix F′ also play an important role in substrate recognition. Helix G is a little far away from the substrate-binding pocket and may not be directly involved in substrate recognition.
The central region of helix I was identified as the SRS4 site in other P450s. In the AOS structure complexed with 13(S)-HODE, a basic residue (Lys-282) in the C-terminal portion of helix I is located near the entrance of channel 1 and forms a charge–charge interaction with the carboxyl group of the substrate analog (Fig. 4A). Lys-282 may play important roles in substrate recognition and binding. Asn-276 pointing to the heme may interact with the 13-hydroperoxyl group if the true substrate is present in the same position (Figs. 4A and 5), suggesting a critical role in catalysis. The unique I helix in AOS is very important for AOS function, especially its signature sequence region (F275–K282) that is involved in heme binding, substrate access, and catalysis.
The loop after helix K and the N-terminal portion of strand β1–4 are regarded as the SRS5 site in other P450s. This region in AOS contains four prolines and one valine and is close to the short ω-chain of 13(S)-HODE with a distance of ≈4 Å from Val-344 and Pro-346. This region is also close to the heme pyrrole rings A and D.
A long loop linking β3–2 and β3–3 is present at the entrance of the active site of AOS. The corresponding region in other P450s is usually even longer, may be present as a small β-sheet 4 that protrudes into the active site and has been regarded as SRS6. In AOS, this loop has a deletion of 14 residues and is nearly located on the molecular surface. Two hydrophobic residues, Pro-458 and Leu-459, point to the long fatty acid chain of 13(S)-HODE, providing a hydrophobic environment. The temperature factor of this loop is very high in all AOS structures, and the loop is nearly disordered in the Native 2 crystal form, suggesting flexibility in this loop region and a possible minor conformational change during substrate binding and product release. This distinctive loop in AOS, together with the loop F/F′, defines the entrances for substrates to access the active site through the channels described above.
The C-terminal region of helix A′ and the N-terminal region of helix A as well as the loop between helices A′ and A are part of the entrance to channel 2. A basic residue (Arg-36) on helix A is present at the entrance. This residue or region may also be involved in substrate recognition and/or product release.
All of these SRSs are highly conserved among AOSs, CYP74A, and CYP74C families (Fig. S3), except for rice (Oryza sativa) AOS, which has five fewer residues in the SRS6 site and may have a more open entrance. These SRSs are quite different in other P450s, including bacterial and mammalian P450s (Fig. S1), and they are highly variable in both sequences and structures, indicating their distinct substrate specificities and functions.
Implications for Substrate Specificity and Catalytic Mechanism.
In the structure of AOS complex with substrate analog, the hydroxyl group at the C13 position of 13(S)-HODE is ≈7 Å from the heme iron. The substrate 13(S)-HPOD may be situated in a similar position, but should be closer to the heme iron. In a model with the substrate replaced in the active site, the substrate hydroperoxyl group is ≈2.4 Å from the heme iron (Fig. 5). Asn-276 in the active site near the heme may interact through a hydrogen bond with the substrate's hydroperoxyl group on position C13. Ser-110 forms a hydrogen bond with Asn-276. This Asn-276 and the related hydrogen network may play an important role in catalysis and substrate binding.
The basic residue Lys-282 located in the center of helix I and at the entrance of the active site may form a charge–charge interaction with the carboxyl group in the long α-end of the fatty acid substrate and is likely a key residue for substrate binding and recognition. Ser-199 forms a hydrogen bond with Lys-282 and may also interact with the substrate and play a role in substrate binding. To explore the roles of these residues in catalysis and substrate binding, mutagenesis studies of AOS will be necessary.
It has been proposed that the position of the hydroperoxyl group in relation to the methyl end is the key for substrate specificity in AOS (11). In the structure of AOS complexed with substrate analog, the methyl end of 13(S)-HODE is extended into the channel 2 and surrounded by residues in the SRS1 (i.e., F92–T93), SRS2 (V206, T209–L210), SRS4 (Ile-283), SRS5 (P343–P346), and SRS6 (P458–V459) (Fig. 4). It is also close to Tyr-29 and Arg-36 in the A′ and A helices. These interactions are mainly hydrophobic but very broad and extensive, indicating the complexity of substrate specificity. 9-AOS and 9/13-AOS in the CYP74C subfamily may also recognize 9(S)-hydroperoxides as substrates. With 9(S)-hydroperoxide placed in the active site of AOS, the methyl end would be extended closely to the residues at the entrance of channel 2, such as Arg-36. For the 9- and 9/13-AOS, an alanine residue is present in the position corresponding to Arg-36 in AOS and may contact or be close to the methyl end of the 9(S)-hydroperoxide.
Similar to other fatty acid hydroperoxide rearrangements, a homolytic cleavage of the hydroperoxide would be the first step in the conversion of 13(S)-hydroperoxide catalyzed by AOS (37). 13(S)-Hydroperoxide binds to the heme iron via the C13 oxygen atom first, and residues Asn-276 and Lys-282 assist the binding and recognition by interacting with the substrate's hydroperoxyl group and carboxyl group, respectively (Fig. 5). Heme-assisted homolytic scission of the oxygen–oxygen bond gives an alkoxyl radical and an iron–oxo complex. The alkoxyl radical then undergoes cyclization with the C11–C12 double bond, to yield a C11 radical, which is then oxidized by the iron–oxo complex to form a carbocation. Loss of a proton from the carbocation finally gives the allene oxide.
Cytochrome P450s catalyze an enormously wide range of chemical reactions and have different enzymatic mechanisms and complex substrate specificities. AOS and other CYP74 enzymes recognize specific fatty acid hydroperoxides in the synthesis of oxygenated lipids. These crystal structures provide a basis for exploring the interactions between enzyme and substrates for deciphering the catalytic mechanism of AOS and the related P450s, and reveal insights into their structure–function relationship.
Methods
Protein Expression and Purification.
The coding region of the AOS cDNA from guayule (P. argentatum) (7) was cloned into the pET28b expression vector with a hexahistidine tag included in the C terminus of the protein. Protein was expressed in Escherichia coli BL21(DE3) cells and purified with Ni2+-nitrilotriacetic acid agarose and a HiprepSephacryl S 200HR gel filtration column according to the procedures reported in ref. 33.
Selenomethionine (SeMet)-substituted AOS was prepared by expressing the recombinant protein in E. coli B834 (DE3) cells (Novagen) grown in M9 minimal medium supplemented with l-SeMet (Sigma) and purified by following the same protocol used for the native protein.
Crystallization and Data Collection.
Crystallization of the native AOS was carried out by using the hanging-drop vapor diffusion method, as described in ref. 33. Briefly, two forms of crystals were obtained from 0.2 M (NH4)H2PO4, 50% 2-methyl-2,4-pentanediol, 0.1 M Tris (pH 8.5). The tetragonal crystal form (Native 1) belongs to the space group I422 with cell parameters a = b = 126.5, c = 163.9 Å, α = β = γ = 90°. A 2.4-Å native dataset from a tetragonal form of crystal was collected.
Because AOS contains a heavy atom Fe, i.e., the heme iron, a native AOS crystal was used to collect Fe anomalous data by using a wavelength of 1.742 Å with an ADSC Quantum 315 CCD detector at the SBC 19ID beamline of the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL). A SeMet derivative crystal was used to collect Se anomalous data at the Southeast Regional Collaborative Access Team (SER-CAT) 22ID beamline in the APS. Because of crystal decay, only one useful SeMet dataset was collected at a wavelength of 0.972 Å.
Crystals of AOS complexed with substrate analog were obtained by cocrystallization with 0.1 mM 13(S)-HODE (Sigma) under the same crystallization conditions used for native protein crystals. The same crystal form with a similar unit cell was obtained, and a 2.6-Å diffraction dataset was collected with a Raxis IV++ image plate detector and RU3H x-ray source.
Another native crystal form (Native 2) with a small unit cell (a = b = 113.5 Å, c = 163.8 Å, α = β = γ = 90°) was obtained by dehydration treatment. A high-resolution dataset at 1.8 Å from this crystal form was collected at the SER-CAT 22ID beamline in the APS. All datasets were processed by using the program suite HKL2000 (38).
Structure Determination and Refinement.
The structure of AOS was determined by using the MIRAS method. Besides the Se and Fe anomalous data, collected at wavelengths of 0.972 Å and 1.742 Å, the 2.4-Å data collected from a native crystal by using the RU3H x-ray source at a wavelength of 1.5418 Å was used as another Fe anomalous dataset. The program SHARP (39) was used to locate and refine sites of the Se and Fe atoms, yielding an overall figure of merit of 0.22. The phase calculation and density modification were also carried out by using SHARP. Autotracing with the program ARP/wARP (40) was performed and generated a model containing 377 aa corresponding to 80% of the protein molecule. Further interactive model building and crystallographic refinement were carried out with a native dataset at 2.4 Å by using the programs COOT (41) and CNS (42), respectively.
The structure of the complex of AOS with the substrate analog 13(S)-HODE were determined at 2.6 Å by the difference Fourier method by using the native structure determined above as a template.
The structure in the Native 2 crystal form with small-unit cell was determined at 1.8 Å by the molecular replacement method with the program PHASER (43), with the structure determined above as a search model.
B factors were refined individually for two AOS native structures at 1.8 and 2.4 Å, and grouped B factor refinement was performed for the structure of AOS complexed with 13(S)-HODE at 2.6 Å. Water molecules were added with ARP/wARP (40) and checked manually for inclusion. In the models, the first five amino acid residues in the N terminus were not observed in AOS–analog structure and the native 1 structure. The last enzyme residue and eight residues including the hexahistidine tag introduced by cloning into the C terminus were disordered in all structures. The geometries of these structures were checked by the program PROCHECK (44). All backbone φ–ψ torsion angles are within allowed regions of the Ramachandran plot.
Supplementary Material
Acknowledgments.
We thank Drs. Y. Tang and H. Pan for critical reading of the manuscript, and K. Tan at the Structural Biology Center beamline 19ID at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL) for assistance with data collection. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. This work was supported by the Samuel Roberts Noble Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3DAM, 3DAN, and 3DBM).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804099105/DCSupplemental.
References
- 1.Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Werck-Reichhart D, Feyereisen R. Cytochromes P450: A success story. Genome Biol 1 reviews. 2000;3003:1–9. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blee E. Impact of phyto-oxylipins in plant defense. Trends Plants Sci. 2002;7:315–322. doi: 10.1016/s1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- 4.Pohnert G. Diatom/copepod interactions in plankton: The indirect chemical defense of unicellular algae. Chembiochem. 2005;6:1–14. doi: 10.1002/cbic.200400348. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann E, Zerbe P, Schaller F. The crystal structure of Arabidopsis thaliana allene oxide cyclase: Insights into the oxylipin cyclization reaction. Plant Cell. 2006;18:3201–3217. doi: 10.1105/tpc.106.043984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skrzypczak-Jankun E, Amzel LM, Kroa BA, Funk MO., Jr Structure of soybean lipoxygenase L3 and a comparison with its L1 isoenzyme. Proteins. 1997;29:15–31. [PubMed] [Google Scholar]
- 7.Pan Z, et al. The major protein of guayule rubber particles is a cytochrome P450. J Biol Chem. 1995;270:8487–8494. doi: 10.1074/jbc.270.15.8487. [DOI] [PubMed] [Google Scholar]
- 8.Song W-C, Funk CD, Brash AR. Molecular cloning of an allene oxide synthase: A cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA. 1993;90:8519–8523. doi: 10.1073/pnas.90.18.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song W-C, Brash AR. Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science. 1991;253:781–784. doi: 10.1126/science.1876834. [DOI] [PubMed] [Google Scholar]
- 10.Hughes RK, et al. Allene oxide synthase from Arabidopsis thaliana (CYP74A1) exhibits dual specificity that is regulated by monomer–micelle association. FEBS Lett. 2006;580:4188–4194. doi: 10.1016/j.febslet.2006.06.075. [DOI] [PubMed] [Google Scholar]
- 11.Stumpe M, Feussner I. Formation of oxylipins by CYP74 enzymes. Phytochem Rev. 2006;5:347–357. [Google Scholar]
- 12.Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C. Allene oxide synthases of barley (Hordeum vulgare cv. Salome): Tissue-specific regulation in seedling development. Plant J. 2000;21:199–213. doi: 10.1046/j.1365-313x.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- 13.Itoh A, Schilmiller AL, McCaig BC, Howe GA. Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. J Biol Chem. 2002;277:46051–46058. doi: 10.1074/jbc.M207234200. [DOI] [PubMed] [Google Scholar]
- 14.Stumpe M, et al. Identification of an allene oxide synthase (CYP74C) that leads to formation of a-ketols from 9-hydroperoxides of linoleic and linolenic acid in below-ground organs of potato. Plant J. 2006;47:883–896. doi: 10.1111/j.1365-313X.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Poulos TL. The structure of the cytochrome P450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat Struct Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 16.Poulos TL, Finzel BC, Howard AJ. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 17.Cupp-Vickery JR, Poulos TL. Structure of cytochrome P450eryF involved in erythromycin biosynthesis. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 18.Hasemann CA, Ravichandran KG, Peterson JA, Deisenhofer J. Crystal structure and refinement of cytochrome P450terp at 2.3 Å resolution. J Mol Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 19.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson JA, Deisenhofer J. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450s. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 21.Yano JK, et al. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J Biol Chem. 2000;275:31086–31092. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 22.Yan JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 23.Scott EE, et al. An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams PA, et al. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 25.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: Structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 26.Yano JK, et al. The structure of human microsomal cytochrome P450 3A4 determined by x-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 27.Schoch GA, et al. Structure of human microsomal cytochrome P450 2C8: Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 28.Rowland P, et al. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 29.Williams PA, et al. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 30.Park SY, et al. Crystal structure of nitric oxide reductase from denitrifying fungus Fusarium oxysporum. Nat Struct Biol. 1997;4:827–832. doi: 10.1038/nsb1097-827. [DOI] [PubMed] [Google Scholar]
- 31.Chiang CW, Yeh HC, Wang LH, Chan NL. Crystal structure of the human prostacyclin synthase. J Mol Biol. 2006;364:266–274. doi: 10.1016/j.jmb.2006.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YC, et al. Structures of prostacyclin synthase and its complexes with substrate analog and inhibitor reveal a ligand-specific heme conformation change. J Biol Chem. 2008;283:2917–2926. doi: 10.1074/jbc.M707470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang Z, Li L, Pan Z, Wang X. Crystallization and preliminary x-ray analysis of allene oxide synthase, cytochrome P450 CYP74A2 from Parthenium argentatum. Acta Crystallogr F. 2008;64:668–670. doi: 10.1107/S1744309108017545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 35.Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Probing the open state of cytochrome P450cam with ruthenium-linker substrates. Proc Natl Acad Sci USA. 2001;98:12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 37.Song WC, Baertschi SW, Boeglin WE, Harris TM, Brash AR. Formation of epoxyalcohols by a purified allene oxide synthase. Implications for the mechanism of allene oxide synthesis. J Biol Chem. 1993;268:6293–6298. [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 40.Lamzin VS, Perrakis A, Wilson KS. In: International Tables for Crystallography: Crystallography of Biological Macromolecules. Rossmann MG, Arnold E, editors. Vol F. Dordrecht: Kluwer; 2001. pp. 720–722. [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallograph D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 45.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 46.Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.