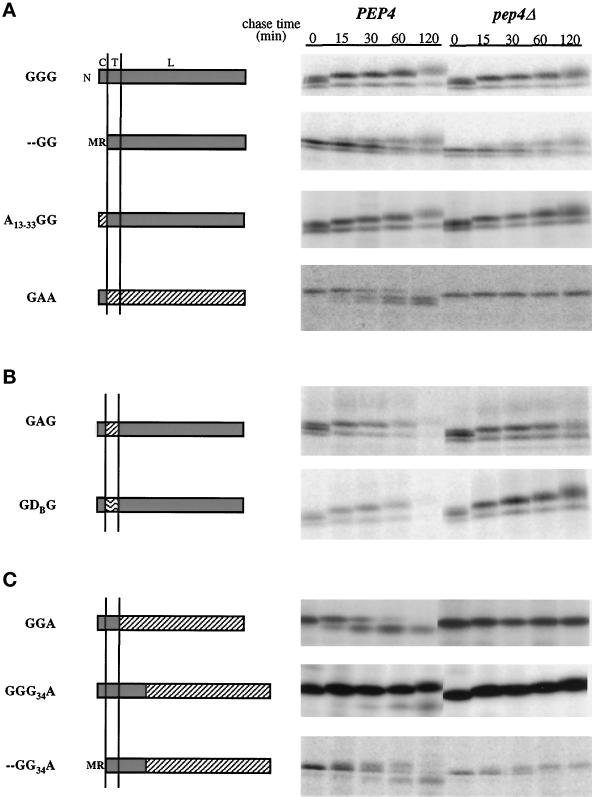
Figure 4.
Pulse–chase analysis of wild-type and mutant GDPase proteins in PEP4 and pep4Δ cells. Proteins are named from amino (N) to carboxy terminus, and letters refer to the protein of origin, where G represents GDPase, A represents ALP, and D represents DPAP B. The domain structure of each protein is diagrammed with GDPase domains shaded, ALP domains diagonally hatched, and DPAP B domains wave hatched. Above the GGG diagram, C refers to the cytoplasmic domain, T to the transmembrane domain, and L to the lumenal domain. Vertical lines represent the membrane. Proteins GGG, - -GG, A13–33GG, GAG, and GDG were expressed and analyzed in strains G2-11 (gda1Δ PEP4) and GPY1452 (gda1Δ pep4Δ), and proteins were precipitated with antibodies to GDPase. Chimeric molecules GAA, GGA, GGG34A, and - -GG34A were expressed and analyzed in strains DKY6280 (pho8Δ PEP4) and GPY1250 (pho8Δ pep4Δ), and antibodies to ALP were used to precipitate the proteins of interest. Labelings and immunoprecipitations were as described for Figure 2A. (A) Wild-type GDPase and mutant and chimeric proteins used to analyze the role of the GDPase cytoplasmic domain in Golgi localization. (B) Chimeric proteins used to test the role of the GDPase transmembrane domain in Golgi localization. (C) Chimeric GDPase proteins used to analyze the role of the lumenal domain in GDPase localization.