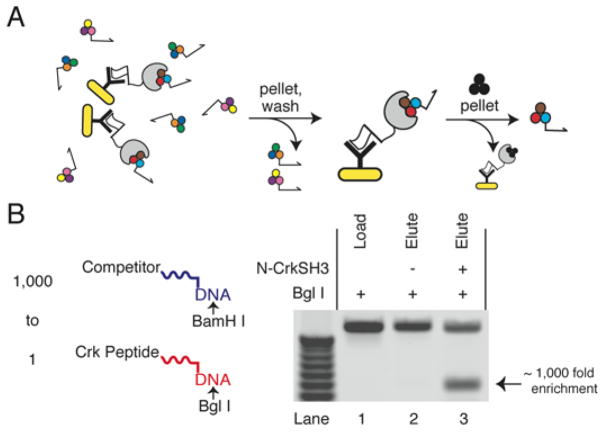
Figure 3.
Selection for N CrkSH3 binding. (A) The selection strategy utilizes an N-CrkSH3 protein (grey) with a prepended FLAG tag (white), as well as an anti FLAG antibody (black Y), and Pansorbin cells (yellow). After incubation of these components with a translated library, the Pansorbin cells are pelleted and washed. Ligand DNA conjugates are eluted by addition of an excess of an N-CrkSH3 binding peptide (black balls). (B) To measure the selection signal lo noise ratio, two peptide DNA conjugates were synthesized. One construct consisted of the known N-CrKSH3 peptide ligand YPPPALPPKRRR linked to a 340 mer dsDNA with a central BgII site (red). The other consisted of the peptide YGGFL linked to a 340 mer dsDNA with a central BamHI site (blue). When the ligand construct was diluted 1 to 1,000 into the non-ligand construct and amplified, BgII cutting was undetectable (lane 1). When this mixture was subjected to a mock selection that lacked N-CrkSH3 and was amplified, BgII cutting was undetectable (lane 2). When subjected to a selection with N-CrkSH3 included and amplified, the BgII containing DNA was enriched nearly 1,000 fold (lane 3).