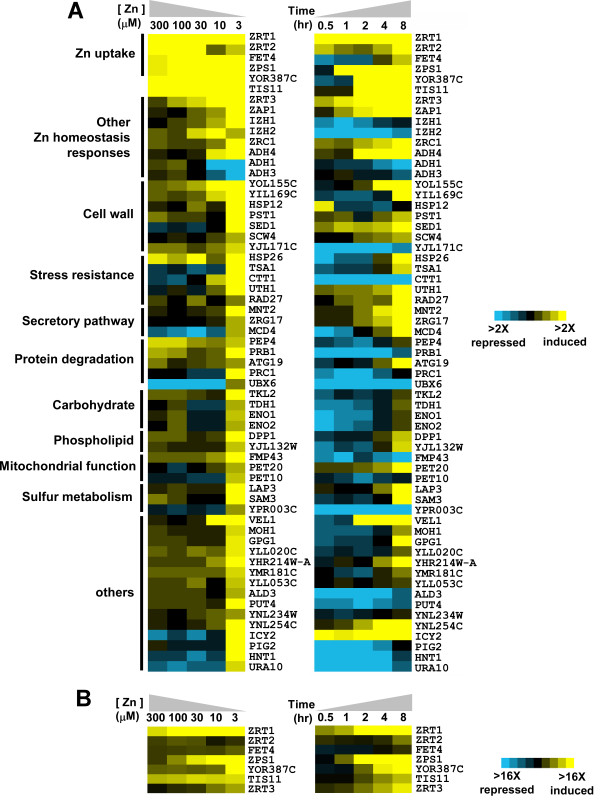
Figure 4.
Differential regulation of Zap1 target genes in response to zinc. A) Microarray studies were performed with cells grown over a range of zinc (left panel) or over time after zinc withdrawal (right panel). For the dose-response analysis, cells were grown in LZM + 3 mM ZnCl2, or LZM + 300, 100, 30, 10 or 3 μM ZnCl2. Transcript levels were assayed using microarrays in which each sample was paired with the zinc-replete (LZM + 3 mM ZnCl2) control. For the time-course studies, cells were grown to exponential phase in a zinc-replete medium (LZM + 1 mM ZnCl2) and then transferred to a zinc-limiting medium (LZM + 1 μM ZnCl2) for 8 hours. RNA was isolated at the indicated times and transcript levels were assayed using microarrays in which each sample was paired with the zinc-replete To control. Genes were grouped by related function and the results are displayed using the Java Treeview program http://jtreeview.sourceforge.net. A narrow color intensity scale (yellow, increased expression relative to control; blue decreased expression) is used to show the conditions under which changes in gene expression were first detectable. B) The data for highly expressed genes in panel A were plotted with a broader scale to better show the differences in gene expression. The complete dose-response and time-course analyses were performed twice with similar results and the data, presented as the ratio relative to control, are provided in Additional file 5.