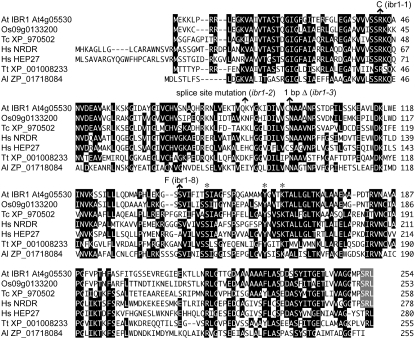
Figure 5.—
IBR1 resembles short-chain dehydrogenase/reductase enzymes. An alignment comparing IBR1 to similar proteins from rice (Os), beetle (Tc; Tribolium castaneum), human (Hs), Tetrahymena (Tt), and the prokaryote Algoriphagus sp. PR1 (Al) was generated in the MegAlign program (DNAStar) using the Clustal W method. Amino acid residues identical in at least four sequences are against a solid background; hyphens indicate gaps introduced to maximize alignment. The ibr1 mutations are indicated above the sequence. The C-terminal PTS1 present in IBR1 and the three closest homologs is indicated by a shaded background; HsHEP27 is not targeted to peroxisomes (Pellegrini et al. 2002). Asterisks above the sequence denote three essential residues that compose the catalytic triad characteristic of SDR proteins (Kallberg et al. 2002).