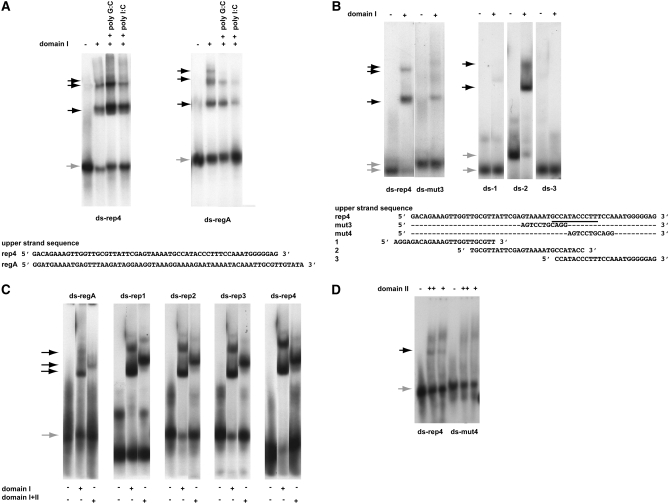
Figure 3.—
DNA-binding specificities of the two Rtf1 c-myb-like domains. A key above the panels defines the experimental conditions used. Unbound (shaded arrows) and shifted material (solid arrows) are indicated for each panel. Unless otherwise stated, the experiments were done in the presence of 100 μg/ml poly G:C. The DNA olignucleotides utilized are given at the bottom of each panel. Western analysis of shifted material is provided for A and B as supplemental data (Figure S2F). (A) Gel-shift assays using purified domain I protein and a dsDNA oligonucleotide resembling motif 4 (ds-rep4; left) and region A (ds-regA; right). The sequences of the “upper” strands of the dsDNA oligonucleotides are displayed. In both panels, DNA binding challenged by the addition of 50 μg/ml nonspecific competitors (given) does not abolish the observed shifts. However, binding can be efficiently outcompeted by the addition of specific competitors constituted by unlabeled substrates (supplemental Figure S2G). (B) Definition of the domain I's binding site within the motif 4 sequence. (Left) Three gel-shift assays using three different segments of motif 4. The sequences of the upper strands of the three dsDNA oligonucleotides, 1, 2, and 3, are displayed. (Right) Gel-shift assay utilizing dsDNA oligonucleotides ds-rep4 or ds-mut3. The different mobilities observed for unbound wild-type and mutant dsDNA oligonucleotides are due to the presence of a GATC overhang on the ds-mut3 oligonucleotide. (C) Comparison of domain I's and the chimeric domain's affinities to the five different dsDNA oligonucleotides constituting region A (ds-regA) and each of the four repeated region B motifs (ds-rep1, 2, 3, and 4). Unbound and shifted material is indicated to the left of the panels with shaded and solid arrows, respectively. The names of the utilized oligonucleotides are shown above the panels. It should be noted, that the retardation observed using the chimeric domains is greater than that observed for the individual domains. Since the increased retardation reflects the increased molecular size the observation establishes independently that the gel-shifts are due to binding of the purified domain(s). (D) Domain II interacts weakly with motif 4. Gel-shift assay using purified domain II protein and dsDNA oligonucleotides ds-rep4 and ds-mut4. A weak gel-shift is only observed with the wild-type sequence (ds-rep4) but not the mutant (ds-mut4). Importantly, we do not see this shift in the presence of unspecific poly I:C competitor DNA (data not shown), suggesting that while the result obtained using the ds-mut4 oligo indicates that the interaction is sequence specific the interaction must be weak as it can be outcompeted with an unspecific competitor. Also, the smear observed in the top section of lanes 2, 3, 5, and 6 is due to the domain II interacting with single-stranded oligo DNA (see below).