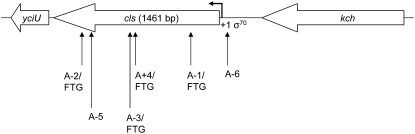
Figure 2.—
Schematic of mutations in cls. Block arrows indicate the direction of transcription for each gene and are drawn to scale. Upward pointing arrows indicate the location of IS150 (A+4/FTG, A-1/FTG, A-3/FTG) and IS186 (A-5, A-6) insertions and an 11-bp deletion (A-2/FTG) in FTG-evolved clones or progenitors already harboring insertions. See supplemental Table 5 for information on the location of each mutation and the number of mutated clones in each population. The bent arrow indicates the putative transcription start site of cls, and the sigma factor (σ70) responsible for its transcription is also shown (Ivanisevic et al. 1995). The yciU gene (330 bp) encodes a hypothetical protein and is predicted to be transcribed in the same direction as cls. In this case, the cls mutations may have polar effects on yciU transcription. The kch gene (1254 bp) encodes a potassium voltage-gated ion channel.