Abstract
As a fitness trait, survival is assumed to exhibit low heritability due to strong selection eroding genetic variation and/or spatio-temporal variation in mortality agents reducing genetic and increasing residual variation. The latter phenomenon in particular may contribute to low heritability in multigeneration data, even if certain cohorts exhibit significant genetic variation. Analysis of survival data from 10 year classes of rainbow trout reared at three test stations showed that treating survival as a single trait across all generations resulted in low heritability (h2 = 0.08–0.17). However, when heritabilities were estimated from homogeneous generation and test station-specific cohorts, a wide range of heritability values was revealed (h2 = 0.04–0.71). Of 64 genetic correlations between different cohorts, 20 were positive, but 16 were significantly negative, confirming that genetic architecture of survival is not stable across generations and environments. These results reveal the existence of hidden genetic variation for survival and demonstrate that treating survival as one trait over several generations may not reveal its true genetic architecture. Negative genetic correlations between cohorts indicate that overall survival has limited potential to predict general resistance, and care should be taken when using it as selection criterion.
FITNESS traits such as survival and fecundity are commonly assumed to have low or close to zero heritability (Fisher 1930; Mousseau and Roff 1987; Roff and Mousseau 1987; Price and Schluter 1991; Merilä and Sheldon 1999; Coltman et al. 2005). Genetic analyses of survival have typically supported this assumption by finding little evidence of genetic variation from diverse taxonomic groups both in farmed (e.g., Van Arendonk et al. 1996; Knol et al. 2002; Goyache et al. 2003; Casellas et al. 2007) and in wild animals (e.g., Futuyma et al. 1995; Campbell 1997). However, in some instances moderate to high heritabilities for survival have also been found (e.g., Robison and Luempert 1984; Ernande et al. 2004; Kenway et al. 2006). Even with fairly low heritability of survival, animal breeding programs have produced slow but significant long-term genetic improvement in survival, contributing to increased animal welfare and economic profitability (Knol et al. 2002; Goyache et al. 2003).
The low heritability of survival is generally hypothesized to be a result of strong selection on this important fitness component, reducing additive genetic variance for survival (Fisher 1930; Mousseau and Roff 1987; Roff and Mousseau 1987). Additionally, it is not evident that multiple agents of mortality (e.g., different diseases, predators, physiological effects) share common genetic determination, thus possibly lowering survival heritability by reducing genetic variance and/or increasing residual variance (Price and Schluter 1991; Houle 1992; Hoffmann and Merilä 1999; Merilä and Sheldon 1999). The latter phenomenon in particular may occur when heritability of survival is estimated across large multigeneration data sets. Parents and offspring may experience different mortality factors that have clear heritable effects, resulting in moderate survival heritability within both cohorts. However, if the mortality factors do not share a common genetic basis, there will be a low parent–offspring regression that will contribute to a low heritability across the whole multigeneration data, even if there is heritable variation within both cohorts. This creates a situation where significant genetic variation existing within specific cohorts is hidden. This possibility, however, has remained unexplored.
For survival, an example of such hidden genetic variation could be a situation, where during certain years mortality is caused by a single major factor (e.g., a specific disease) for which there exists high genetic variation, while during other years, there may be multiple mortality factors and heritability is therefore low. It is well established that different environments affect heritability of traits (Hoffmann and Merilä 1999; Kause and Morin 2001; Charmantier and Garant 2005). Moreover, different cohorts (e.g., generations, herds, test stations) may experience different mortality factors that each display significant genetic variation, but the factors may be weakly or even negatively correlated with each other. Resistance to different mortality factors would then display genetic trade-offs (Gjøen et al. 1997; Cotter et al. 2004; Henryon et al. 2005; Luong and Polak 2007). In such a case, heritability from the whole combined data set may produce a zero-heritability estimate, even when cohort factors are correctly modeled in the statistical analysis.
To test for the existence of hidden genetic variation for survival, we analyzed extensive data on survival of rainbow trout (Oncorhynchus mykiss Walbaum) collected from 10 year classes of the Finnish national breeding program. The benefit of using rainbow trout to test our hypotheses is that due to the semiwild production environment, trout are exposed to natural variability in climate conditions, diseases, and parasites that create extensive spatial and generation-to-generation variability in mortality factors. Fish breeding programs also have discrete generations and distinct production environments. For instance, in addition to the abiotic environment differences, freshwater and sea have diseases that are specific to each environment. This differs from terrestrial farm animals that are often held under more standardized environmental conditions, and distinguishing separate generations and effects of different environments is more difficult. In wild animals, multigeneration pedigreed populations have only recently become available for genetic analysis (Quinn et al. 2006; Kruuk and Hill 2008) but recording of survival remains a challenge.
In this study, we first estimated heritability of survival in two environments across the whole data set, to find out whether or not the assumption of low heritability for overall survival is fulfilled. Second, to assess whether or not the genetic architecture of survival remains stable through space and time, the whole data were split into separate generations and three test stations, and cohort-specific heritabilities as well as genetic correlations between survival in different cohorts were estimated.
METHODS
The data originated from the Finnish national rainbow trout breeding program maintained by the Finnish Game and Fisheries Research Institute and MTT Agrifood Research Finland at the Tervo freshwater nucleus station in Central Finland (Kause et al. 2005).
Population structure:
The data included 121,905 individuals with observations originating from 10 year classes from 1995 to 2004 reared at one to three test stations located either in freshwater or in the sea (Table 1). A total of 1159 ancestors without observations and fertilized in 1989, 1990, 1992, and 1993 were included to complete the pedigree. The fish belonged to three subpopulations (PopI, PopIIa, and PopIIb; Table 1) with three to four successive generations. Each year class consisted of 109–341 full-sib families generated from 48–168 sires and 79–252 dams, mated using either nested paternal or partial factorial designs. The total numbers of fish within each year class were 4459–13,643 in the freshwater station and 1456–5165 in the sea stations (Tables 1 and 2). All populations share a common base population from which parents were sampled in 1989 for PopI and in 1990 for PopIIa and PopIIb. PopIIa and PopIIb diverged after one generation, with generations in 1996 and 1997 sharing six sires and 23 dams born in 1993. Population structure is detailed in Table 1.
TABLE 1.
Population structure and mating designs
| Population/generation | Fertilization year | Sires | Dams | Mean (range) dams per sire | Mean (range) sires per dam | Full-sib families | Family tanks | Sea test stations |
|---|---|---|---|---|---|---|---|---|
| Population I | ||||||||
| 1 | 1995 | 92 | 272 | 3.0 (1–5) | 1.0 (1–1) | 272 | 370 | — |
| 2 | 1998 | 71 | 128 | 1.8 (1–4) | 1.0 (1–1) | 128 | 132 | — |
| 3 | 2001 | 121 | 154 | 2.5 (1–6) | 2.0 (1–3) | 303 | 303 | 2 |
| 4 | 2004 | 130 | 93 | 1.9 (1–5) | 2.7 (1–4) | 250 | 250 | 2 |
| Population IIa | ||||||||
| 1 | 1996 | 75 | 150 | 2.0 (1–4) | 1.0 (1–1) | 150 | 150 | — |
| 2 | 1999 | 48 | 109 | 2.3 (1–4) | 1.0 (1–1) | 109 | 150 | 2 |
| 3 | 2002 | 113 | 139 | 2.5 (1–6) | 2.1 (1–3) | 287 | 287 | 1 |
| Population IIb | ||||||||
| 1 | 1997 | 65 | 79 | 2.9 (1–5) | 2.4 (1–3) | 191 | 228 | — |
| 2 | 2000 | 98 | 122 | 2.0 (1–5) | 1.6 (1–3) | 200 | 200 | 2 |
| 3 | 2003 | 168 | 155 | 2.0 (1–5) | 2.2 (1–3) | 341 | 341 | 2 |
TABLE 2.
Trait abbreviations, sample sizes, heritabilities (h2), common environment effects (c2) and their standard errors (SE), genetic (VG) and phenotypic (VP) variances estimated across the whole freshwater or sea data (OverallF and OverallS, respectively) and in cohort-specific split data (G95T-G03B)
| Population/fertilization year | Locationa | Trait | Sample size | Survival (%) | c2 ± SE | h2 ± SE | VG | VP |
|---|---|---|---|---|---|---|---|---|
| All | F | OverallF | 81499 | 72 | 0.05 ± 0.00 | 0.17 ± 0.02 | 0.0258 | 0.1975 |
| All | Sa + Sb | OverallS | 40405 | 71 | 0.04 ± 0.00 | 0.08 ± 0.02 | 0.0156 | 0.1934 |
| PopI | ||||||||
| 1995 | F | G95F | 13643 | 80 | 0.04 ± 0.01 | 0.22 ± 0.05 | 0.0176 | 0.1634 |
| 1998 | F | G98F | 6096 | 74 | 0.03 ± 0.02 | 0.37 ± 0.11 | 0.0399 | 0.1962 |
| 2001 | F | G01F | 8399 | 50 | 0.02 ± 0.01 | 0.19 ± 0.04 | 0.0296 | 0.2504 |
| 2001 | Sa | G01Sa | 3929 | 58 | 0.00 ± 0.01 | 0.10 ± 0.03 | 0.0147 | 0.2451 |
| 2001 | Sb | G01Sb | 3948 | 54 | 0.03 ± 0.01 | 0.12 ± 0.05 | 0.0190 | 0.2495 |
| 2004 | F | G04F | 7501 | 71 | 0.00 ± 0.01 | 0.10 ± 0.03 | 0.0115 | 0.2076 |
| 2004 | Sa | G04Sa | 3748 | 79 | 0.01 ± 0.01 | 0.07 ± 0.04 | 0.0059 | 0.1662 |
| 2004 | Sb | G04Sb | 3745 | 88 | 0.02 ± 0.01 | 0.04 ± 0.05 | 0.0017 | 0.1050 |
| PopIIa | ||||||||
| 1996 | F | G96F | 7480 | 78 | 0.01 ± 0.01 | 0.20 ± 0.06 | 0.0172 | 0.1712 |
| 1999 | F | G99F | 4459 | 74 | 0.00 ± 0.01 | 0.71 ± 0.11 | 0.0819 | 0.2121 |
| 1999 | Sa | G99Sa | 1863 | 68 | 0.01 ± 0.02 | 0.68 ± 0.13 | 0.0980 | 0.2463 |
| 1999 | Sb | G99Sb | 1770 | 71 | 0.04 ± 0.02 | 0.32 ± 0.10 | 0.0395 | 0.2184 |
| 2002 | F | G02F | 5503 | 65 | 0.06 ± 0.02 | 0.24 ± 0.07 | 0.0331 | 0.2280 |
| 2002 | Sa | G02Sa | 1456 | 60 | 0.00 ± 0.02 | 0.27 ± 0.10 | 0.0414 | 0.2447 |
| PopIIb | ||||||||
| 1997 | F | G97F | 10262 | 73 | 0.04 ± 0.01 | 0.18 ± 0.05 | 0.0172 | 0.1683 |
| 2000 | F | G00F | 7916 | 77 | 0.04 ± 0.01 | 0.38 ± 0.08 | 0.0378 | 0.1901 |
| 2000 | Sa | G00Sa | 4932 | 80 | 0.04 ± 0.01 | 0.34 ± 0.08 | 0.0268 | 0.1642 |
| 2000 | Sb | G00Sb | 4798 | 75 | 0.01 ± 0.01 | 0.45 ± 0.08 | 0.0479 | 0.1983 |
| 2003 | F | G03F | 10240 | 71 | 0.03 ± 0.01 | 0.35 ± 0.05 | 0.0438 | 0.2178 |
| 2003 | Sa | G03Sa | 5051 | 56 | 0.02 ± 0.01 | 0.20 ± 0.05 | 0.0320 | 0.2509 |
| 2003 | Sb | G03Sb | 5165 | 84 | 0.05 ± 0.01 | 0.70 ± 0.11 | 0.0454 | 0.1453 |
Values significantly different from zero are in italics.
F, Tervo freshwater nucleus station; Sa, Sb, sea test stations.
Rearing procedures:
The parents for each generation were selected on the basis of their estimated breeding values for growth (since 1992), maturity age (2001), external appearance (2001), fillet color (2003), and cataract by Diplostomum parasite (2003) (Kause et al. 2005). Parental fish were mated at the Tervo freshwater nucleus station during April–June. Full-sib egg batches were incubated separately, and at the eyed-egg stage, each full-sib family was transferred to one or two indoor 150-liter family tanks (Table 1). Eggs hatched in July. During the winter after hatching, fingerlings were removed from the family tanks and individually tagged with passive integrated transponders (Trovan, Germany).
After tagging, the fish were transferred to an outdoor raceway at the freshwater station and/or sent to one or two Baltic Sea test stations (Table 1). At the freshwater station the fish were held in a flow-through earth-bottomed raceway. All sea stations were located in southwest Finland within a maximum distance of 163 km from each other, but they were not the same ones from generation to generation. At the sea stations, the fish were reared under commercial farming conditions in a net pen. All fish were fed commercial fish feed pellets throughout the rearing cycle. The rearing procedure is detailed by Kause et al. (2005).
Trait definitions:
The survival data were collected after the tagged fish had spent one growing season either at the freshwater or at the sea station. At the end of the growing season, all fish remaining were captured and their individual tags recorded. The fish that survived from tagging to the end of the growing season were scored as survived (= 0), while fish not present in the end were coded as missing (= 1).
To estimate heritability over all generations, a trait “overall survival” was defined as survival at the freshwater station (trait OverallF, Table 2) and survival at the sea stations (trait OverallS, Table 2) across all generations. To estimate cohort-specific genetic architecture, the data were next split into 21 separate survival traits, by defining survival in each generation and in each of the three test stations as separate traits (Table 2).
Genetic analysis:
Heritabilities and genetic correlations between cohorts for survival were estimated using restricted maximum-likelihood and multitrait animal models (DMU-AI software; Madsen and Jensen 2008).
The model for overall survival at the freshwater station was
 |
(1) |
and for overall survival at the sea stations was
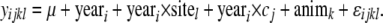 |
(2) |
The 21 generation- and test-stationwise traits were modeled as
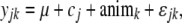 |
(3) |
where μ is a mean of a given trait, yeari is the fixed effect of fertilization year (i = 10 years), yeari × cj is the random interaction effect of birth year with common environment effect shared by full sibs before tagging (j = number of family tanks), yeari × sitel is the fixed interaction effect of birth year and sea test station (l = 2 sites), animk is the random genetic animal effect (k = number of animals), and ɛ is a random error term. The common environment effect was modeled separately for each birth year because each tank does not have a consistent effect every year.
Both overall survival traits were analyzed simultaneously in a single two-trait analysis using models 1 and 2, respectively. These models estimate genetic (co)variation of the traits in the whole population because all animals originated from the same base population. Heritabilities and genetic correlations for generation- and test-stationwise analysis were derived from one multitrait model (using model 3) run performed separately for each population, resulting in eight, six, and seven trait runs for PopI, PopIIa, and PopIIb, respectively. Because within a population, all cohort- and generation-specific traits were included in a single multitrait run, the potential effect of selection bias on variance components should be reduced (Ouweltjes et al. 1988).
Heritability was quantified as h2 = VG(VG + VC + VR)−1, where VG is genetic variation, VC variation due to common environment, and VR residual variation. Although genetic variance is assumed to be mainly due to additive genetic effects, the potential effects of dominance and epistasis cannot, however, be excluded. The common environment effect was quantified as c2 = VC(VC + VG + VR)−1. In addition to common environment effects of full sibs, VC may potentially include parts of dominance variance. Asymptotic standard errors for genetic parameters were computed on the basis of a Taylor series approximation (Madsen and Jensen 2008).
Heritabilities estimated by the linear model were transformed to the underlying liability scale using the formula of Dempster and Lerner (1950). Genetic correlations of binary traits estimated using linear models are unbiased (Mäntysaari et al. 1991). Because only one survival trait was recorded from each individual, residual covariance was set to zero when calculating all the correlations. Moreover, when calculating genetic correlations across generations, covariance due to the common environment was set to zero.
Genetic covariance between survival in different generations was assessed in two ways. First, genetic correlations between generations were estimated. Second, for a subset of the cohorts, a correlation was calculated between the breeding values of parents estimated either from the observations of their own generation or from those of their offspring's generation only (Ødegård et al. 2007). The latter can be regarded as realized breeding values of the parents. A correlation between the two types of breeding values was calculated for the parental individuals contributing to the offspring generation (n = 144–364 parents in each generation). The model for estimating breeding values was the same as the above used for the split data (model 3).
Furthermore, to enhance the visualization of genetic correlation matrices, a principal component analysis was conducted (Rao 1964). The principal components for each population were calculated separately from their estimated full genetic correlation matrices. The analysis (PROC PRINCOMP) was conducted using SAS v. 9.1.3 (Sas 2005).
RESULTS
Overall survival—whole data:
As expected, heritability for overall survival across all generations and subpopulations was low (Table 2). In the sea, heritability was low but significantly different from zero (h2 = 0.08 ± 0.02). At the freshwater station, heritability was 0.17 ± 0.02 (Table 2). Genetic correlation between freshwater and sea was positive and significantly different from both zero and unity (rG = 0.58, 95% confidence interval = 0.39–0.76), indicating moderate genotype–environment interaction.
Cohortwise survival—split data:
Heritabilities within cohorts:
When all generations and test stations were analyzed as separate traits, an extensive variability in the heritability values was revealed, the 21 individual heritabilities ranging from 0.04 to 0.71 (Table 2). The highest heritabilities were in the generations 1999, 2000, and 2003. The means of the individual heritabilities (0.29 and 0.30 at freshwater and sea stations, respectively) were higher than the heritabilities for overall survival.
Genetic correlations between generations:
Of the 46 genetic correlations across generations, 8 were significantly positive (range 0.39–0.89), 16 significantly negative (range −0.97– −0.45), and 22 did not differ from zero (range −0.54–0.54; Table 3). Similar to the variation in heritabilities, this suggests again that the genetic architecture of survival does vary between generations. In addition to being correlations across generations, some of these correlations are also between test stations in different generations.
TABLE 3.
Genetic correlations and their standard errors between generations and test stations in three subpopulations
| G95F | G98F | G01F | G01Sa | G01Sb | G04F | G04Sa | |
|---|---|---|---|---|---|---|---|
| PopI | |||||||
| G98F | 0.48 ± 0.27 | ||||||
| G01F | 0.21 ± 0.43 | 0.52 ± 0.19 | |||||
| G01Sa | −0.20 ± 0.56 | 0.02 ± 0.28 | 0.29 ± 0.16 | ||||
| G01Sb | −0.38 ± 0.68 | 0.24 ± 0.31 | 0.39 ± 0.17 | 0.90 ± 0.19 | |||
| G04F | 0.47 ± 0.90 | 0.22 ± 0.42 | 0.13 ± 0.24 | 0.73 ± 0.27 | 0.48 ± 0.32 | ||
| G04Sa | −0.48 ± 1.04 | 0.13 ± 0.51 | −0.15 ± 0.31 | 0.70 ± 0.36 | 0.82 ± 0.40 | 0.38 ± 0.24 | |
| G04Sb | −0.34 ± 1.36 | −0.53 ± 0.72 | −0.54 ± 0.47 | 0.19 ± 0.54 | 0.07 ± 0.58 | 0.17 ± 0.33 | 0.37 ± 0.46 |
| G96F | G99F | G99Sa | G99Sb | G02F | |||
| PopIIa | |||||||
| G99F | −0.83 ± 0.18 | ||||||
| G99Sa | −0.72 ± 0.22 | 0.97 ± 0.04 | |||||
| G99Sb | −0.74 ± 0.28 | 0.95 ± 0.09 | 0.94 ± 0.11 | ||||
| G02F | 0.54 ± 0.40 | −0.45 ± 0.22 | −0.40 ± 0.24 | −0.41 ± 0.30 | |||
| G02Sa | 0.89 ± 0.41 | −0.91 ± 0.18 | −0.85 ± 0.24 | −0.90 ± 0.32 | 0.71 ± 0.19 | ||
| G97F | G00F | G00Sa | G00Sb | G03F | G03Sa | ||
| PopIIb | |||||||
| G00F | 0.49 ± 0.24 | ||||||
| G00Sa | 0.61 ± 0.24 | 0.93 ± 0.07 | |||||
| G00Sb | 0.49 ± 0.23 | 0.89 ± 0.06 | 0.98 ± 0.05 | ||||
| G03F | −0.51 ± 0.30 | −0.84 ± 0.11 | −0.96 ± 0.11 | −0.97 ± 0.08 | |||
| G03Sa | −0.03 ± 0.36 | −0.69 ± 0.17 | −0.58 ± 0.20 | −0.61 ± 0.17 | 0.68 ± 0.10 | ||
| G03Sb | −0.20 ± 0.32 | −0.75 ± 0.13 | −0.86 ± 0.13 | −0.93 ± 0.10 | 0.94 ± 0.05 | 0.73 ± 0.09 | |
Trait definitions are in Table 2. Correlations significantly different from zero in are in italics (95% confidence intervals do not include zero).
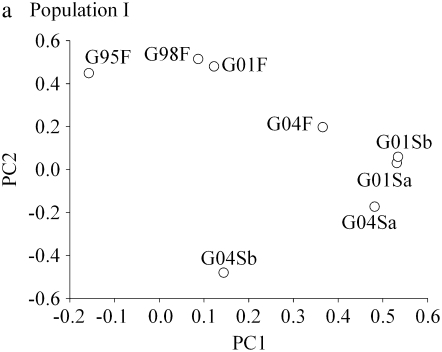
When the results are examined within each population, clear patterns emerge for the negative correlations (Table 3). In PopI, genetic correlations between generations at the freshwater station were positive (mean correlation = 0.34, range 0.13–0.52), whereas the negative correlations occurred only between the freshwater station and the sea stations (mean correlation between generations = −0.08, range −0.54–0.73). This result was also clearly visualized by the results of the principal component analysis on the full genetic correlation table for this population (Figure 1a). When the first two principal components were plotted, the freshwater traits were located in the top left-hand corner, whereas the seawater traits were located in the bottom right-hand corner. That two principal components were needed to clearly separate fresh- and seawater environments is logical because each of the two first principal components explained 30–40% of the variation (Table 4).
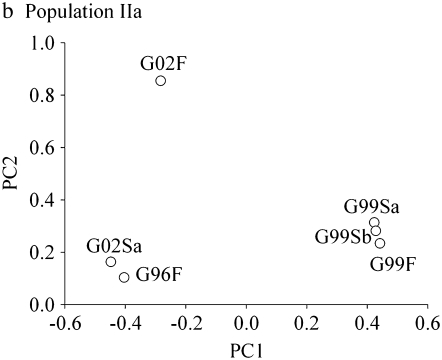
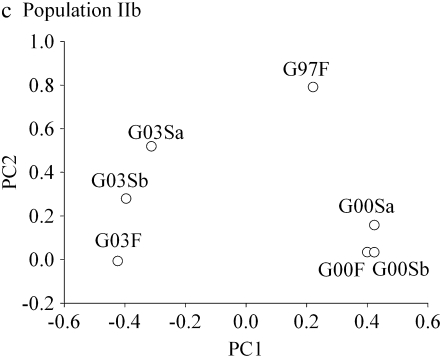
Figure 1.—
Individual survival traits plotted along the first and second principal component axes for each population. Trait abbreviations are given in Table 2.
TABLE 4.
Principal component analysis of the full genetic correlation matrices for the three populations
| PC | Eigenvalue | Prop% | Cum% |
|---|---|---|---|
| PopI | |||
| 1 | 3.22 | 40.3 | 40.3 |
| 2 | 2.47 | 30.9 | 71.2 |
| 3 | 1.29 | 16.1 | 87.3 |
| 4 | 0.68 | 8.6 | 95.9 |
| PopIIa | |||
| 1 | 4.82 | 80.3 | 80.3 |
| 2 | 0.80 | 13.3 | 93.6 |
| 3 | 0.29 | 4.9 | 98.5 |
| 4 | 0.08 | 1.3 | 99.8 |
| PopIIb | |||
| 1 | 5.37 | 76.7 | 76.7 |
| 2 | 1.08 | 15.4 | 92.1 |
| 3 | 0.36 | 5.1 | 97.2 |
| 4 | 0.19 | 2.7 | 99.9 |
For each of the first four principal components (PC), eigenvalues, proportion of variance explained (Prop%), and cumulative explanation percentage (Cum%) are given.
The patterns of both PopIIa and PopIIb differed from that of PopI, but do resemble each other. Both PopIIa and PopIIb were characterized by a single generation that is negatively correlated with the other generations. In PopIIa, the genetic correlations between 1996 and 2002 were positive [rG (SE): G96F vs. G02F, 0.54 (0.40); G96F vs. G02Sa, 0.89 (0.41)], but these generations were negatively correlated with 1999 (mean genetic correlation = −0.69, range −0.91– −0.40; Table 3). The first principal component explained 80.3% of the variance (Table 4) and clearly separated generation 1999 from the two others (Figure 1b).
Similarly to the pattern found in PopIIa, in PopIIb the generation 2003 correlated negatively with generations 1997 and 2000 (mean correlation = −0.66, range −0.91– −0.40; Table 3) and the generations 1997 and 2000 were significantly positively correlated with each other (range 0.49–0.61; Table 3). Again most of the variation was explained by the first principal component (76.7%) and in the plot of the first and second principal components the generation 2003 formed its own cluster (Table 4, Figure 1c).
Correlations between breeding values calculated from parental and offspring generations:
In general, correlations of breeding values produced results similar to the genetic correlations between generations, revealing both significant negative and positive correlations (Table 5). Of 18 correlations for breeding values, 6 were significantly positive, 5 significantly negative, and 7 nonsignificantly different from zero.
TABLE 5.
Comparison of genetic correlations (rG) with correlations between breeding values estimated from parental and offspring generation observations (rS)
| Parent trait | Offspring trait | rG | rS |
|---|---|---|---|
| PopI | |||
| G95F | G98F | 0.48 | 0.04 |
| G98F | G01F | 0.52 | 0.44 |
| G98F | G01Sa | 0.02 | 0.17 |
| G98F | G01Sb | 0.24 | 0.34 |
| G01F | G04F | 0.13 | 0.08 |
| G01F | G04Sa | −0.15 | −0.08 |
| G01F | G04Sb | −0.54 | −0.13 |
| PopIIa | |||
| G96F | G99F | −0.83 | −0.25 |
| G96F | G99Sa | −0.72 | −0.19 |
| G96F | G99Sb | −0.74 | −0.13 |
| G99F | G02F | −0.45 | −0.39 |
| G99F | G02Sa | −0.91 | −0.24 |
| PopIIb | |||
| G97F | G00F | 0.49 | 0.14 |
| G97F | G00Sa | 0.61 | 0.36 |
| G97F | G00Sb | 0.49 | −0.11 |
| G00F | G03F | −0.84 | 0.07 |
| G00F | G03Sa | −0.69 | −0.05 |
| G00F | G03Sb | −0.75 | 0.14 |
Trait definitions are in Table 2. rG, genetic correlation between two cohorts; values with their standard errors are shown in Table 3. rS, Spearman's correlation between breeding values estimated from parental generation observations in the freshwater station and their next generation progeny reared at three test stations. Values significantly different from zero are in italics.
In PopI and PopIIa, genetic correlations and correlations between breeding values were the same sign and produced similar patterns. For PopIIb, in two of six cases the genetic correlations were significantly negative but the correlations between breeding values were significantly positive (Table 5). Generally, the two methods produced similar results.
Genetic correlations between test stations within generations:
In general, genetic correlations between the test stations within generations were much more stable and more positive than those between generations (Table 3). Of the 16 genetic correlations calculated between the test stations within generations, 12 were significantly positive (range 0.39–0.98), of which 9 did not differ from unity. Four correlations were positive but not significantly different from zero (range 0.17–0.38; Table 3). In line with the analysis of overall survival across the whole data, these results substantiate that survival exhibits moderate genotype–environment interaction and that the genetic architecture of survival varies from test station to test station.
DISCUSSION
On the basis of our genetic analysis of multigeneration data from the pedigreed population of rainbow trout, we showed that genetic architecture of survival is not stable across generations and environments. Moreover, even negative genetic correlations can occur between survival in distinct cohorts. Spatio-temporal variation in genetic architecture for this closely related fitness trait resulted in low heritability for overall survival across the whole data set, hiding significant genetic variation in specific cohorts.
Revealing hidden genetic variation:
When survival in each of the two environments was regarded as a single trait across all generations, overall survival showed low heritability in both environments (freshwater h2 = 0.17 ± 0.02, sea h2 = 0.08 ± 0.02). While significantly different from zero, these heritability values are still in accordance with the hypothesis of low heritability for fitness-related traits (Fisher 1930; Mousseau and Roff 1987; Roff and Mousseau 1987; Price and Schluter 1991; Merilä and Sheldon 1999; Coltman et al. 2005). For instance, the review by Mousseau and Roff (1987) showed mean heritability of 0.26 for life-history traits assumed to be under strong selection and heritability of 0.46 for morphological traits assumed to be under weaker selection.
Our results confirm the assumption that the low heritability for overall survival may occur because there are different mortality factors in separate cohorts that do not share common genetic determination, thus blurring consistent differences between genotypes across cohorts. However, when our data were analyzed separately for each generation and test station, large variability in heritability values was revealed, the maximum heritability value being 0.71. Consequently, the genetic background of survival was not homogenous across the cohorts, and treating survival as one trait over several generations may hide its true genetic architecture. These statements are highlighted further by the fact that many of the genetic correlations across the generations were low or negative. By splitting the data into homogeneous cohorts, we were able to reveal genetic variation that would otherwise have been hidden. Similarly, in a study of selection response in the beginning of the Nile tilapia (Oreochromis niloticus) breeding program, Charo-Karisa et al. (2006) reported varying heritability of survival for the three generations (range 0.03–0.14). The reader should note that also the differences between the generations in mating ratios and population structure as well as sampling error may produce minor variation in the heritability estimates. However, these factors alone cannot explain the huge variability observed.
Furthermore, it is possible, yet speculative, that selection may have eroded genetic variation from overall survival, even if its component traits show high genetic variation. It is well established that separate components of survival, such as mortality against specific diseases or parasites, may display negative genetic correlations, facilitating the maintenance of genetic variation in the component traits (Gjøen et al. 1997; Cotter et al. 2004; Henryon et al. 2005; Leimu and Koricheva 2006; Ødegård et al. 2007).
Despite the well-justified logic that selection, genetic drift, and mutations modify genetic architecture of traits (Fisher 1930; Kimura 1958; Falconer 1960), early predictive evolutionary quantitative genetic models assumed that genetic architecture of traits remains unchanged (Lande 1979). However, several experimental approaches have since illustrated the way heritabilities and genetic correlations are modified in time and space. First, the impact of selection on genetic architecture has been assessed using artificial selection experiments (Shaw et al. 1995) and by comparing genetics of behavioral, morphological, and life-history traits assumed to be under different selection pressures (Gustafsson 1986; Mousseau and Roff 1987; Roff and Mousseau 1987; Kruuk et al. 2000). Second, the effects of mutation have been studied within laboratory populations (Houle et al. 1996; Camara and Pigliucci 1999). Third, comparative studies have examined the way genetic architecture has evolved at macroevolutionary timescales (Kause et al. 2001; Steppan et al. 2002). Fourth, genetic architecture has been shown to vary with environment quality (Kause and Morin 2001; Sgró and Hoffmann 2004; Charmantier and Garant 2005; Roff and Fairbairn 2007). Fifth, it has been shown that trait variation depends on its position in a trait hierarchy, where high-level life-history traits may accumulate genetic and/or environmental variation of the lower-level morphological and behavioral traits influencing them (Price and Schluter 1991; Houle 1992; Kause et al. 1999). Accordingly, currently the question is under which conditions, and why, does the genetic architecture evolve and vary (Steppan et al. 2002).
Our study is most closely related to the fourth and fifth categories. However, the originality of our study is that the variability of genetic characteristics of survival depends on trait expression, i.e., variation in factors causing mortality in the first place. Survival differs from conventional traits due to its complex origin. Especially crucial is that survival is influenced by underlying component traits whose expression may vary in time and space. Interestingly, it can be further envisioned that the magnitude of the genetic effect of a mortality factor may depend on the type and number of previous mortality factors. The correlation between them in particular will determine whether the new factor will magnify or cancel out the genetic variation induced by the previous factors. Although survival is defined and measured similarly throughout the study, it is a product of multiple (and mostly unknown) factors that may vary in their incidence between environments and generations. It is logical to assume that this phenomenon is also applicable to other traits with similar multicausal determination yet with a simple trait definition. Indeed, our findings of heterogeneity in genetic (co)variances across generations substantiate results by Kause et al. (2007), who found that the heritability of rainbow trout deformations is elevated only during the years when deformations are found in uncommonly high incidence and that these generations are negatively genetically correlated with other generations.
Is survival a measure of general resistance?
For animal breeders, a common goal is to select animals that have general resistance or robustness against multiple environmental disturbances, stressors, and mortality factors. This topic is currently becoming a major target of research because it strongly contributes to increased animal welfare and ability to manage animals across a wide range of environments (Mulder and Bijma 2005; Pertoldi et al. 2007). Likewise, in the wild, robustness influences the fitness of individuals, making understanding of robustness genetics and its developmental mechanisms of importance for evolutionary biology (Félix and Wagner 2008). With the advent of advanced molecular tools (Schwartz et al. 2007), it is now possible to assess and monitor genetic parameters of survival and other previously logistically unfeasible traits also in wild animal populations.
When survival data are collected over a large number of generations, overall survival is a potential easy measure of general resistance, given that genetic correlations between different cohorts are positive. In our analysis, many of the genetic correlations between generations and test stations were positive, but some specific cohorts displayed strong negative genetic correlations with the other cohorts. The negative correlations occurred systematically with certain generations (1999 and 2003 in PopIIa and PopIIb, respectively) or between freshwater and sea stations (PopI). Negative genetic correlations are genetic trade-offs, where the genetic background increasing survival in the presence of certain mortality factor(s) may reduce survival in the presence of another factor(s). Genetic trade-offs between resistance to different diseases have indeed been found in rainbow trout (Henryon et al. 2005) and Atlantic salmon (Gjøen et al. 1997).
These results imply that overall survival is not a perfect measure of general resistance, and care should be taken when using it as a selection criterion. In the worst-case scenario, resistance to a single factor may be reduced when selection is based on overall survival or survival in a single abnormal year. This may even lead to maladaptations in certain environments. An example of general resistance is presented in the meta-analysis by Leimu and Koricheva (2006), who found that genetic correlations between plant resistance to multiple natural enemies were mainly positive. Similarly, inbred lines or clones of aphids (Ferrari et al. 2001) and Daphnia (Decaestecker et al. 2003) have been shown to be resistant to several parasite, parasitoid, or pathogen species. In humans it has been suggested that some genotypes are less susceptible to multiple disorders (Rzhetsky et al. 2007).
The genetic correlations between freshwater and sea environments were positive in the overall analysis as well as in the cohortwise analyses. This suggests that to some extent the same genetic architecture is responsible for survival in different environments, in which different mortality factors may prevail. In contrast to the between-generation correlations, within generations there were no negative genetic correlations between the test stations. This may be understandable because within a generation, all paternal and maternal families experience similar initial conditions, including initial growth and potential diseases, which is likely to make correlations between environments positive rather than negative within a generation. For example, if the main mortality factor during a certain year is an infectious disease that has already attacked the fish during fingerling growth, family differences against this disease will create positive genetic correlations across all the test stations when mortality occurs during grow-out period. Such shared initial events do not influence genetic correlations calculated across generations.
In this study, principal components were used to visualize the patterns of the large correlation tables. As suggested by a reviewer, also direct estimates of genetic principal components could have been estimated from the raw data (Kirkpatrick and Meyer 2004; Meyer and Kirkpatrick 2005; Hine and Blows 2006). Using this approach, it is possible to identify the number of underlying latent variables and their genetic variation, generating the observed multiple genetic correlations (e.g., Blows et al. 2004; Mezey and Houle 2005). This would provide an additional vigorous way to quantify the degree of genetic variation for general resistance and robustness.
In conclusion, the results have two important implications. First, survival, a low-heritability trait closely related to fitness, displayed both zero and very high heritabilities when the data were split to more homogeneous cohorts. This implies that spatio-temporal variation may hide the underlying genetic architecture of overall survival. Second, overall survival across generations did not accurately measure general resistance or robustness, because some specific generations displayed strong negative genetic correlations with the other cohorts. Our study highlights the unique genetic characteristics that survival can have.
Acknowledgments
This study would not have been possible to conduct without the extensive data set collected by hard-working Finnish Game and Fisheries Research Institute staff at the Tervo station. Martin Lidauer, Esa Mäntysaari, Timo Serenius, and Ismo Strandén, as well as other members of Biometrical Genetics, Animal Genomics, and Genetic Diversity at MTT Agrifood Research Finland, and two reviewers willingly shared their vast knowledge of genetics and methodology in numerous occasions. This study was financially supported by the Finnish Ministry of Agriculture and Forestry and the Kone Foundation.
References
- Blows, M. W., S. F. Chenoweth and E. Hine, 2004. Orientation of the genetic variance-covariance matrix and the fitness surface for multiple male sexually selected traits. Am. Nat. 163 329–340. [DOI] [PubMed] [Google Scholar]
- Camara, M., and M. Pigliucci, 1999. Mutational contributions to genetic variance/covariance matrices; an experimental approach using induced mutations in Arabidopsis thaliana. Evolution 53 1692–1703. [DOI] [PubMed] [Google Scholar]
- Campbell, D. R., 1997. Genetic and environmental variation in life-history traits of a monocarpic perennial: a decade-long field experiment. Evolution 51 373–382. [DOI] [PubMed] [Google Scholar]
- Casellas, J., G. Caja, X. Such and J. Piedrafita, 2007. Survival analysis from birth to slaughter of Ripollesa lambs under semi-intensive management. J. Anim. Sci. 85 512–517. [DOI] [PubMed] [Google Scholar]
- Charmantier, A., and D. Garant, 2005. Environmental quality and evolutionary potential: lessons from wild populations. Proc. R. Soc. Ser. B 272 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charo-Karisa, H., H. Komen, M. A. Rezk, R. W. Ponzoni, J. A. M. Van Arendonk et al., 2006. Heritability estimates and response to selection for growth of Nile tilapia (Oreochromis niloticus) in low-input earthen ponds. Aquaculture 261 479–486. [Google Scholar]
- Coltman, D. W., P. O'Donoghue, J. T. Hogg and M. Festa-Bianchet, 2005. Selection and genetic (co)variance in bighorn sheep. Evolution 59 1372–1382. [PubMed] [Google Scholar]
- Cotter, S. C., L. E. B. Kruuk and K. Wilson, 2004. Costs of resistance: genetic correlations and potential trade-offs in an insect immune system. J. Evol. Biol. 17 421–429. [DOI] [PubMed] [Google Scholar]
- Decaestecker, E., A. Vergote, D. Ebert and L. De Meester, 2003. Evidence for strong host clone-parasite species interactions in the Daphnia microparasite system. Evolution 57 784–792. [DOI] [PubMed] [Google Scholar]
- Dempster, E. R., and I. M. Lerner, 1950. Heritability of threshold characters. Genetics 35 212–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernande, B., P. Boudry, J. Clobert and J. Haure, 2004. Plasticity in resource allocation based life history traits in the Pacific oyster, Crassostrea gigas. I. Spatial variation in food abundance. J. Evol. Biol. 17 342–356. [DOI] [PubMed] [Google Scholar]
- Falconer, D., 1960. Introduction to Quantitative Genetics. Robert MacLehose & Co., Glasgow.
- Félix, M. A., and A. Wagner, 2008. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity 100 132–140. [DOI] [PubMed] [Google Scholar]
- Ferrari, J., C. B. Muller, A. R. Kraaijeveld and H. C. J. Godfray, 2001. Clonal variation and covariation in aphid resistance to parasitoids and a pathogen. Evolution 55 1805–1814. [DOI] [PubMed] [Google Scholar]
- Fisher, R., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Futuyma, D. J., M. C. Keese and D. J. Funk, 1995. Genetic constraints on macroevolution - the evolution of host affiliation in the leaf beetle genus Ophraella. Evolution 49 797–809. [DOI] [PubMed] [Google Scholar]
- Gjøen, H. M., T. Refstie, O. Ulla and B. Gjerde, 1997. Genetic correlations between survival of Atlantic salmon in challenge and field tests. Aquaculture 158 277–288. [Google Scholar]
- Goyache, F., J. P. Gutierrez, I. Alvarez, I. Fernandez, L. J. Royo et al., 2003. Genetic analysis of calf survival at different preweaning ages in beef cattle. Livest. Prod. Sci. 83 13–20. [Google Scholar]
- Gustafsson, L., 1986. Lifetime reproductive success and heritability—empirical support for Fisher's fundamental theorem. Am. Nat. 128 761–764. [Google Scholar]
- Henryon, M., P. Berg, N. J. Olesen, T. E. Kjaer, W. J. Slierendrecht et al., 2005. Selective breeding provides an approach to increase resistance of rainbow trout (Oncorhynchus mykiss) to the diseases, enteric redmouth disease, rainbow trout fry syndrome, and viral haemorrhagic septicaemia. Aquaculture 250 621–636. [Google Scholar]
- Hine, E., and M. W. Blows, 2006. Determining the effective dimensionality of the genetic variance-covariance matrix. Genetics 173 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A., and J. Merilä, 1999. Heritable variation and evolution under favourable and unfavourable conditions. Trends Ecol. Evol. 14 96–101. [DOI] [PubMed] [Google Scholar]
- Houle, D., 1992. Comparing evolvability and variability of quantitative traits. Genetics 130 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle, D., B. Morikawa and M. Lynch, 1996. Comparing mutational variabilities. Genetics 143 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kause, A., and J. P. Morin, 2001. Seasonality and genetic architecture of development time and body size of the birch feeding sawfly Priophorus pallipes. Genet. Res. 78 31–40. [DOI] [PubMed] [Google Scholar]
- Kause, A., I. Saloniemi, E. Haukioja and S. Hanhimäki, 1999. How to become large quickly: quantitative genetics of growth and foraging in a flush feeding lepidopteran larva. J. Evol. Biol. 12 471–482. [Google Scholar]
- Kause, A., I. Saloniemi, J. P. Morin, E. Haukioja, S. Hanhimäki et al., 2001. Seasonally varying diet quality and the quantitative genetics of development time and body size in birch feeding insects. Evolution 55 1992–2001. [DOI] [PubMed] [Google Scholar]
- Kause, A., O. Ritola, T. Paananen, H. Wahlroos and E. A. Mäntysaari, 2005. Genetic trends in growth, sexual maturity and skeletal deformations, and rate of inbreeding in a breeding programme for rainbow trout (Oncorhynchus mykiss). Aquaculture 247 177–187. [Google Scholar]
- Kause, A., O. Ritola and T. Paananen, 2007. Changes in the expression of genetic characteristics across cohorts in skeletal deformations of farmed salmonids. Genet. Sel. Evol. 37 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenway, M., M. Macbeth, M. Salmon, C. Mcphee, J. Benzie et al., 2006. Heritability and genetic correlations of growth and survival in black tiger prawn Penaeus monodon reared in tanks. Aquaculture 259 138–145. [Google Scholar]
- Kimura, M., 1958. On the change of population fitness by natural selection. Heredity 12 145–167. [Google Scholar]
- Kirkpatrick, M., and K. Meyer, 2004. Direct estimation of genetic principal components: simplified analysis of complex phenotypes. Genetics 168 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knol, E. F., J. I. Leenhouwers and T. Van Der Lende, 2002. Genetic aspects of piglet survival. Livest. Prod. Sci. 78 47–55. [Google Scholar]
- Kruuk, L. E. B., and W. G. Hill, 2008. Introduction. Evolutionary dynamics of wild populations: the use of long-term pedigree data. Proc. R. Soc. Ser. B Biol. Sci. 275 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk, L. E. B., T. H. Clutton-Brock, J. Slate, J. M. Pemberton, S. Brotherstone et al., 2000. Heritability of fitness in a wild mammal population. Proc. Natl. Acad. Sci. USA 97 698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R., 1979. Quantitative genetic analysis of multivariate evolution applied to brain: body size allometry. Evolution 33 402–416. [DOI] [PubMed] [Google Scholar]
- Leimu, R., and J. Koricheva, 2006. A meta-analysis of genetic correlations between plant resistances to multiple enemies. Am. Nat. 168 E15–E37. [DOI] [PubMed] [Google Scholar]
- Luong, L. T., and M. Polak, 2007. Costs of resistance in the Drosophila-macrocheles system: a negative genetic correlation between ectoparasite resistance and reproduction. Evolution 61 1391–1402. [DOI] [PubMed] [Google Scholar]
- Madsen, P., and J. Jensen, 2008. DMU—A Package for Analysing Multivariate Mixed Models, Version 6. www.dmu.agrsci.dk.
- Mäntysaari, E., R. Quaas and Y. Gröhn, 1991. Simulation study on covariance component estimation for two binary traits in an underlying continuous scale. J. Dairy Sci. 74 580–591. [DOI] [PubMed] [Google Scholar]
- Merilä, J., and B. C. Sheldon, 1999. Genetic architecture of fitness and nonfitness traits: empirical patterns and development of ideas. Heredity 83 103–109. [DOI] [PubMed] [Google Scholar]
- Meyer, K., and M. Kirkpatrick, 2005. Restricted maximum likelihood estimation of genetic principal components and smoothed covariance matrices. Genet. Sel. Evol. 37 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezey, J., and D. Houle, 2005. The dimensionality of genetic variation for wing shape in Drosophila melanogaster. Evolution 59 1027–1038. [PubMed] [Google Scholar]
- Mousseau, T. A., and D. A. Roff, 1987. Natural selection and the heritability of fitness components. Heredity 59 181–197. [DOI] [PubMed] [Google Scholar]
- Mulder, H. A., and P. Bijma, 2005. Effects of genotype x environment interaction on genetic gain in breeding programs. J. Anim. Sci. 83 49–61. [DOI] [PubMed] [Google Scholar]
- Ødegård, J., I. Olesen, B. Gjerde and G. Klemetsdal, 2007. Evaluation of statistical models for genetic analysis of challenge-test data on ISA resistance in Atlantic salmon (Salmo salar): prediction of progeny survival. Aquaculture 266 70–76. [Google Scholar]
- Ouweltjes, W., L. R. Schaeffer and B. W. Kennedy, 1988. Sensitivity of methods of variance component estimation to culling type of selection. J. Dairy Sci. 71 773–779. [Google Scholar]
- Pertoldi, C., R. Bijlsma and V. Loeschcke, 2007. Conservation genetics in a globally changing environment: present problems, paradoxes and future challenges. Biodiv. Conserv. 16 4147–4163. [Google Scholar]
- Price, T., and D. Schluter, 1991. On the low heritability of life-history traits. Evolution 45 853–861. [DOI] [PubMed] [Google Scholar]
- Quinn, J. L., A. Charmantier, D. Garant and B. C. Sheldon, 2006. Data depth, data completeness, and their influence on quantitative genetic estimation in two contrasting bird populations. J. Evol. Biol. 19 994–1002. [DOI] [PubMed] [Google Scholar]
- Rao, C., 1964. The use and interpretation of principal component analysis in applied research. Sankhya Ser. A 26 329–358. [Google Scholar]
- Robison, O., and L. Luempert, 1984. Genetic variation in weight and survival of brook trout (Salvelinus fontinalis). Aquaculture 38 155–170. [Google Scholar]
- Roff, D. A., and D. J. Fairbairn, 2007. The evolution of trade-offs: Where are we? J. Evol. Biol. 20 433–447. [DOI] [PubMed] [Google Scholar]
- Roff, D. A., and T. A. Mousseau, 1987. Quantitative genetics and fitness—lessons from Drosophila. Heredity 58 103–118. [DOI] [PubMed] [Google Scholar]
- Rzhetsky, A., D. Wajngurt, N. Park and T. Zheng, 2007. Probing genetic overlap among complex human phenotypes. Proc. Natl. Acad. Sci. USA 104 11694–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS, 2005. SAS/STAT User's Guide, Version 9.1. SAS Institute, Cary, NC.
- Schwartz, M. K., G. Luikart and R. S. Waples, 2007. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22 25–33. [DOI] [PubMed] [Google Scholar]
- Sgró, C. M., and A. A. Hoffmann, 2004. Genetic correlations, tradeoffs and environmental variation. Heredity 93 241–248. [DOI] [PubMed] [Google Scholar]
- Shaw, F. H., R. G. Shaw, G. S. Wilkinson and M. Turelli, 1995. Changes in genetic variances and covariances: G whiz! Evolution 49 1260–1267. [DOI] [PubMed] [Google Scholar]
- Steppan, S. J., P. C. Phillips and D. Houle, 2002. Comparative quantitative genetics: evolution of the G matrix. Trends Ecol. Evol. 17 320–327. [Google Scholar]
- Van Arendonk, J. A. M., C. Van Rosmeulen, L. L. G. Janss and E. F. Knol, 1996. Estimation of direct and maternal genetic (co) variances for survival within litters of piglets. Livest. Prod. Sci. 46 163–171. [Google Scholar]