Abstract
NMR structure determination of helical integral membrane proteins is one of the most challenging undertakings in modern structural biology. The solubilizing detergent micelle or phospholipid bicelle adds significantly to the overall size of the system, often requiring perdeuteration to obtain useable spectra. However, perdeuteration prevents structure determination using traditional NOE analysis. Residual dipolar coupling constraints are an attractive complement to NOE distance constraints, but alignment methods are limited to strained polyacrylamide gels due to the incompatibility of the solubilizing detergent or lipid with alignment media such as bicelles or phage particles. We demonstrate the use of lanthanide ions bound to the protein through a small thiol-linked metal chelator as a robust method for partial alignment of membrane proteins. This method provides multiple alignment orientations depending on the ion bound, and permits RDC measurement of multiple bond vectors. We demonstrate that using this method a large number of RDC's can be measured using 3-dimensional NMR methods where alignment using strained polyacrylamide gels results in fewer peaks due to drastic line-broadening.
While the field of structural biology is advancing rapidly with over 40,000 structures currently in the Protein Data Bank, progress on membrane proteins, which comprise nearly 30% of the human genome, has been much slower; only about 110 of solved structures represent unique membrane proteins. NMR is an attractive alternative to x-ray crystallography for these proteins that are difficult crystallize, and has the added advantage of studying their structure and dynamics in solution.
For membrane protein NMR, the solubilizing detergent micelle or phospholipid bicelle adds significantly to the size of the system, and hence to the rotational correlation time. Thus, the protein behaves as a much larger particle, resulting in weak or missing signals in multidimensional spectra due to rapid R2 relaxation. Deuteration and TROSY1 pulse sequences reduce the experimental relaxation rates permitting resonance assignments, but fully deuterated proteins pose a problem for structure determination using traditional NOE analysis. Residual dipolar coupling orientational constraints2 have seen extensive use in the determination and refinement of deuterated globular protein structures3. However, there are few reports of RDC's being used to define the structures of any membrane proteins by NMR4. Most alignment media – phage, phospholipid bicelles, and organic liquid crystals5 – are incompatible with the detergents or lipids used to solubilize these proteins. Strained polyacrylamide gels are inert and provide different alignment orientations by varying the composition of copolymers6 or the method of straining7. In practice, however, strained gels performed poorly with polytopic helical membrane proteins. With two- and four-helix membrane proteins we have seen poor quality spectra in all tested gels, with weak or missing cross-peaks resulting from the lower achievable protein concentration and restricted tumbling in uncharged gels, and significant interactions between proteins and the matrix in charged gels. Furthermore, neutral acrylamide gels lose alignment over the course of a few days (Figure S1), making the measurement of RDC's by 3D NMR methods impractical. Hence an alternative alignment method was sought that ideally would not reduce the achievable protein concentration, would not significantly increase resonance linewidths, and would yield samples with significant alignment and high quality NMR spectra that were stable for multiple days.
Several reports have described the partial alignment of proteins induced by the binding of lanthanide ions8. Chimeric proteins have been constructed by fusing the target protein to a calcium-binding protein or peptide, such as calmodulin9 or an EF-hand10. A similar approach employed small metal-chelating compounds with thiol-reactive groups, linked to a unique cysteine sidechain in the protein11. Several compounds of varying linker lengths have been synthesized and some are commercially available12. Here we show that the thiol-directed approach overcomes the shortcomings of other alignment media with polytopic helical membrane proteins, providing a robust method for multiple, stable alignments.
Alignment methods were tested using the two transmembrane helix subunit c of the E. coli F1Fo ATP synthase in micelles. An A79C mutation introduced a unique cysteine at the C-terminus of the protein, which should be the most innocuous location for introducing a chemical modification. Modification was achieved by adding 30 μl of 100 mM N-[S-(2-Pyridylthio) cysteaminyl]EDTA (Toronto Research Chemicals, Inc.) to 500 μl of 600 μM subunit c in 5% LPPG (1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)], Avanti Polar Lipids, Alabaster, Al), 100 mM Tris pH 8. The reaction proceeded overnight at 42° C. To incorporate metals, it was necessary to preload the N-[S-(2-Pyridylthio)cysteaminyl]EDTA with the lanthanide ion prior to protein modification, as the free ions caused excessive detergent precipitation. The lanthanides were incorporated by adding a three-fold molar excess of LnCl3 solution to the N-[S-(2-Pyridylthio)cysteaminyl]EDTA sample in the Tris buffer without detergent. After 30 minutes, sufficient EDTA was added to leave a slight (∼5-10%) LnCl3 excess, resulting in negligible precipitation of the NMR sample.
NMR spectra of high quality were obtained for the modified and metal-bound subunit c (Figures 1, 2, & S2). The spectra of the mutant and EDTA-modified mutant were largely unchanged with respect to the wild-type subunit c spectrum. Upon addition of lanthanide ions such as TbCl3, significant pseudocontact shifts were observed for residues 66-78, with smaller shifts for residues 61-65. Some perturbation of the resonances for residues 2, 6, 7, and 8, which are located on the N-terminal helix directly opposite Cys79, were also observed.
Figure 1.
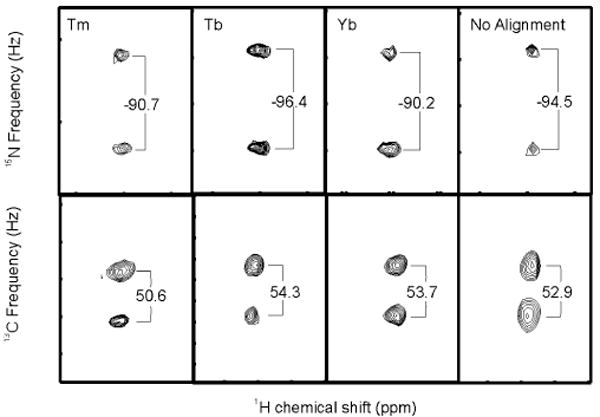
Selected upfield and downfield components of a 3D-IPAP HNCO experiment measuring (A) 1H15N splittings for Gly18 or (B) 13C′13Cα splittings for Phe54. The top panel indicates the lanthanide ion used for alignment or no alignment. Tm+3 and Yb+3 spectra were measured at 800 MHz and Tb was measured at 900 MHz. The observed splitting in Hz is shown.
Figure 2.
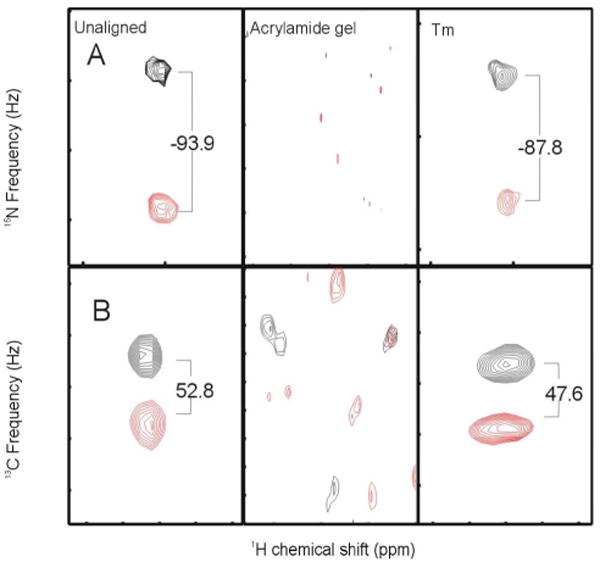
(A) 1H15N and (B) 13C′13Cα splittings for Val15 in an unaligned sample, polyacrylamide gel, or Tm3+ alignment. All spectra were measured at 800 MHz. The observed splittings in Hz are indicated.
Figure 1 shows selected upfield and downfield 1H15N and 13C′13Cα components from a 3D in-phase anti-phase HNCO experiment, with changes in the observed splittings demonstrating lanthanide-induced alignment of the protein in the magnetic field. The alignment tensor depends on the metal used13. Here for example, the Gly18 1H15N RDC was +3.7 Hz with Tm3+ and –2.0 Hz with Tb3+ (Figure 1A), with the opposite sign clearly demonstrating the different alignments with the different metals. Similarly the 13C′-13Cα RDC's for Phe54 were –2.2 Hz with Tm3+ and 1.4 Hz with Tb3+ (Figure 1B). The degree of lanthanide-induced alignment increases with increasing magnetic field strength13; the observed range of 1H15N RDC's for Yb+3 was -6.6 to 5.3 at 800 MHz, and −8.1 to 5.9 at 900 MHz.
Multiple sets of orientational constraints from RDC's will likely be essential for high resolution NMR structures of polytopic helical membrane proteins. Unlike β-barrels, long-range backbone NOE's are vanishingly scarce, and long-range side chain NOE's are difficult to identify unambiguously for these proteins due to extensive chemical shift degeneracy in the side chains. Alignment using strained polyacrylamide gels has yielded useful RDC's for several membrane proteins4, but for helical proteins only measurements for 1H15N vectors using 2D experiments have been reported. The 3D experiments typically required to fully resolve the 1H15N, 13C15N, and 13C′13Cα cross-peaks appear to be impractical in polyacrylamide gels, due to the reduced sample concentrations and restricted rotation in the gels, as well as the lower sensitivity of the 3D experiments themselves. Additionally, relaxation of uncharged gels reduces the usefulness of any measurement that cannot be completed within one or two days. As shown in the middle panels of Figure 2A and 2B, many of the 1H15N and 13C′-13Cα are simply missing from the 3D data sets. Using lanthanide ions to induce alignment, however, we were able to use 3D experiments to readily measure nearly all of the 1H15N and 13C′13Cα RDC's in subunit c (Figure 2A and B, right panels, and Table S1) with multiple orientations induced by varying the identity of the metal.
Protein alignment via lanthanide metals may be crucial for the application of RDC's to solving the NMR structures of polytopic helical membrane proteins such as subunit c. For the partial alignment of such membrane proteins, lanthanide ions proved to be far superior to acrylamide gels. For proteins that naturally contain a cysteine residue, this method is quite simple and easily yields RDC's for full sets of the 1H15N, 13C15N, and 13C′13Cα vectors, with partial alignment that does not decay over time. Multiple alignments are easily obtained by using different metals or different mutations.
Supplementary Material
1H15N spectra of modified and unmodified subunit c in detergent micelles, and tables of RDCs from alignment with Tm3+, Tb3+, and Yb3+. This material is available free of charge at http://pubs.acs.org.
Acknowledgments
This work was supported by NIH grants GM075026 (MG) and GM66354 (NYSBC).
References
- 1.Pervushin K, Riek R, Wider G, Wuthrich K. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tjandra N, Bax A. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 3.(a) Lipsitz R, Tjandra N. Annu Rev Biophys Biomol Struct. 2004;33:413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]; (b) Prestegard J, Bougault C, Kishore A. Chem Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 4.(a) Howell SC, Mesleh MF, Opella SJ. Biochemistry. 2005;44:5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]; (b) Oxenoid K, Chou JJ. Proc Natl Acad Sci U S A. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cierpicki T, Liang B, Tamm LK, Bushweller JH. J Am Chem Soc. 2006;128:6947–6951. doi: 10.1021/ja0608343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prestegard J, Bougault C, Kishore A. Chem Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 6.Cierpicki T, Bushweller JH. J Am Chem Soc. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- 7.(a) Tycko R, Blanco FJ, Ishii Y. J Am Chem Soc. 2000;122:9340–9341. [Google Scholar]; (b) Chou J, Gaemers S, Howder B, Louis J, Bax A. Journal of Biomolecular NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 8.(a) Contreras MA, Ubach J, Millet O, Rizo J, Pons M. J Am Chem Soc. 1999;121:8947–8948. [Google Scholar]; (b) Ma C, Opella SJ. J Magn Reson. 2000;146:381–384. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]; (c) Feeny J, Birdsall B, Bradbury AF, Biekofsky RR, Bayley PM. J Biomol NMR. 2001;21:41–48. doi: 10.1023/a:1011924017938. [DOI] [PubMed] [Google Scholar]; (d) Dvoretsky A, Gaponenko V, Rosevear PR. FEBS Lett. 2002;528:189–192. doi: 10.1016/s0014-5793(02)03297-0. [DOI] [PubMed] [Google Scholar]
- 9.Feeny J, Birdsall B, Bradbury AF, Biekofsky RR, Bayley PM. J Biomol NMR. 2001;21:41–48. doi: 10.1023/a:1011924017938. [DOI] [PubMed] [Google Scholar]
- 10.Ma C, Opella SJ. J Magn Reson. 2000;146:381–384. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]
- 11.(a) Dvoretsky A, Gaponenko V, Rosevear PR. FEBS Lett. 2002;528:189–192. doi: 10.1016/s0014-5793(02)03297-0. [DOI] [PubMed] [Google Scholar]; (b) Pintacuda G, Moshref A, Leonchiks A, Sharipo A, Otting G. J Biomol NMR. 2004;29:351–361. doi: 10.1023/B:JNMR.0000032610.17058.fe. [DOI] [PubMed] [Google Scholar]; (c) Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C. J Biomol NMR. 2004;29:339–349. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]
- 12.Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C. J Biomol NMR. 2004;29:339–349. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]
- 13.Barbieri R, Bertini I, Cavallaro G, Lee YM, Luchinat C, Rosato A. J Am Chem Soc. 2002;124:5581–558. doi: 10.1021/ja025528d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H15N spectra of modified and unmodified subunit c in detergent micelles, and tables of RDCs from alignment with Tm3+, Tb3+, and Yb3+. This material is available free of charge at http://pubs.acs.org.


