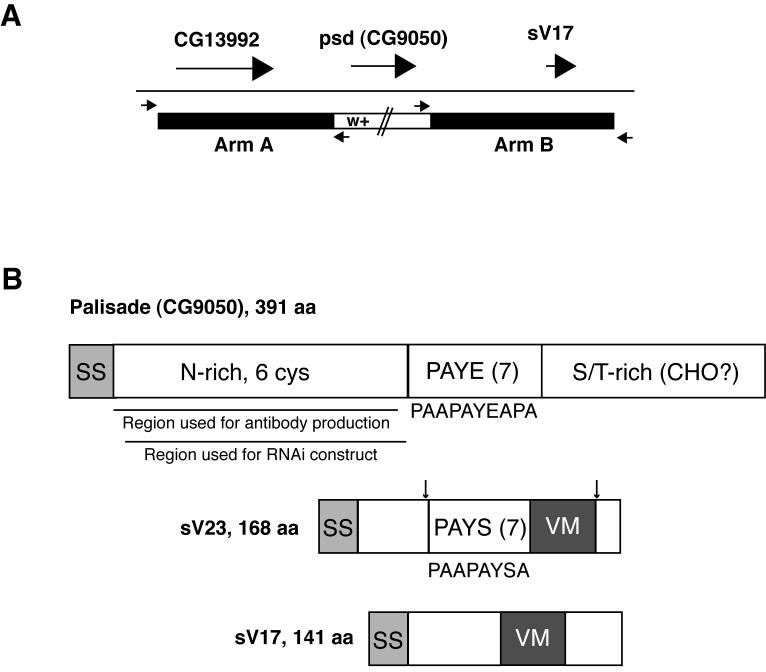
Fig. 1. Palisade gene and protein diagrams.
(A) Illustration of the strategy used for ends-out targeted replacement of the psd (CG9050) gene with the mini-white marker (see Methods). Large arrows represent psd and flanking genes, while small arrows represent primers used to verify the correct insertion of mini-white into the psd locus. (B) Comparison of Palisade protein structure to the sV23 and sV17 major vitelline membrane proteins. (Top) Palisade contains signal sequence (SS), an asparagine-rich domain including 6 of 8 cysteines in the protein (N-rich, 6 cys), a domain containing 7 imperfect copies of a 10 aa repeat (consensus listed below diagram) enriched in proline, alanine, tyrosine, and glutamic acid (PAYE(7)), and a domain enriched in serine and threonine (S/T-rich) and containing 11 of 15 predicted O-glycosylation sites (CHO?). Also shown are regions used for antibody and UAS-psd-RNAi construct generation. (Middle and bottom) sV23 and sV17 share the VM domain, missing in Palisade, while sV23 also contains a repeat region containing 7 copies of an imperfect 8 aa repeat (consensus listed below diagram) enriched in proline, alanine, tyrosine and serine (PAYS(7)). In sV23, approximate positions of N- and C-terminal cleavages are indicated by arrows; sV17 also undergoes cleavage following secretion but sites have not been demonstrated (Manogaran and Waring, 2004; Pascucci et al., 1996).