Fig. 2.
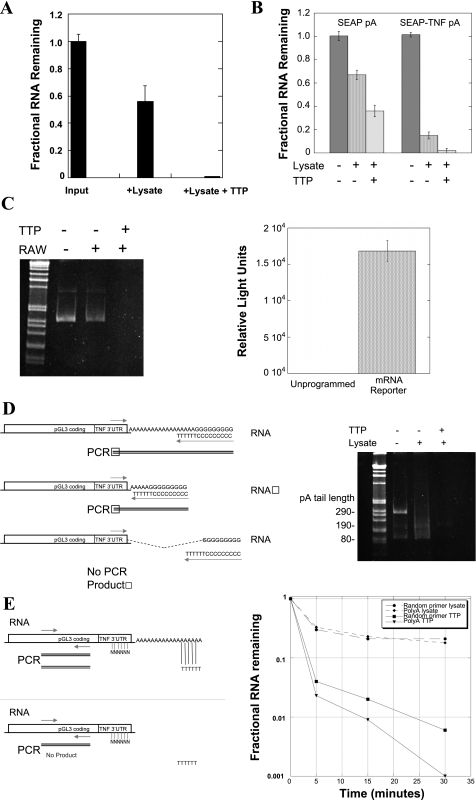
Properties of TTP-promoted deadenylation. RAW264.7 (RAW) cytoplasmic extracts and recombinant TTP were used to create an in vitro system for the study of RNA degradation. Total RNA from RAW264.7 cells stimulated for 1 h with LPS was added with carrier RNA to unstimulated RAW264.7 cell cytoplasmic extracts with or without recombinant TTP for 5 min. The remaining TNF mRNA in each condition was quantified by use of oligo(dT)-primed TaqMan real-time RT-PCR and normalized to the input RNA, shown in A. The role of the TNF 3′-untranslated region (UTR) in this process was confirmed using synthetically polyadenylated pSEAP synthetic RNAs that either lack or include the Balb/c TNF 3′-UTR (SYBRgreen real-time PCR, B). Similar results by use of standard RT-PCR are shown on an agarose gel (C, left) by using a synthetic capped luciferase-TNF 3′-UTR-poly(A)-tailed mRNA and luciferase coding region primers. This capped, polyadenylated mRNA is also transcriptionally active in reticulocyte lysates, but not in our macrophage extracts, confirming that translation is not required for TTP-mediated degradation (C, right). Efforts to measure the rate of deadenylation were unsatisfying due to the rapidity of the TTP-dependent process. One of these, the poly(G) tailing method of Kusov et al. (27), is shown schematically in D, left, which primes reverse transcription and PCR amplification from the extreme end of the poly(A) tail by creating a 3′-primer site at the junction of the 6 terminal A residues and an enzymatically added guanosine tract prior to PCR. Our choice of 5′-primer in the TNF 3′-UTR creates a PCR product whose size is equal to the remaining poly(A) tail length plus 75 bases. The reduction in of poly(A) tail length in RAW cell cytoplasmic extracts (D, center lane on gel) is completely lost when recombinant TTP is added (D, right lane on gel). For subsequent studies, the PCR primers in the luciferase coding region as in C (above) were used so that the efficiency of amplification would be the same in all conditions. The 3′ dependence of the TTP effect was addressed using the luciferase-TNF reporter RNA, but by priming the reverse transcription from either end of the poly(A) tail vs. by random priming from the cRNA body, shown schematically in E, left. Both random oligo-primed and poly(A)-tail primed SYBRgreen real-time RT-PCR were evaluated by use of luciferase primers and the results with and without TTP graphed in E, right, expressed as a fraction of the input RNA at time 0. The same data expressed as the ratio of poly(A)-primed to random-primed RNA from 0–30 min are seen in Table 1, with the ratio between them only changing when recombinant TTP is added. Neomycin, a known inhibitor of several deadenylases such as poly(A) ribonuclease and Ccr4 (36, 40), inhibits the effect of TTP on the reporter RNA (F). All of these experiments were repeated at least 3 times or more, with different TTP, lysate, and RNA preparations.