Figure 3.
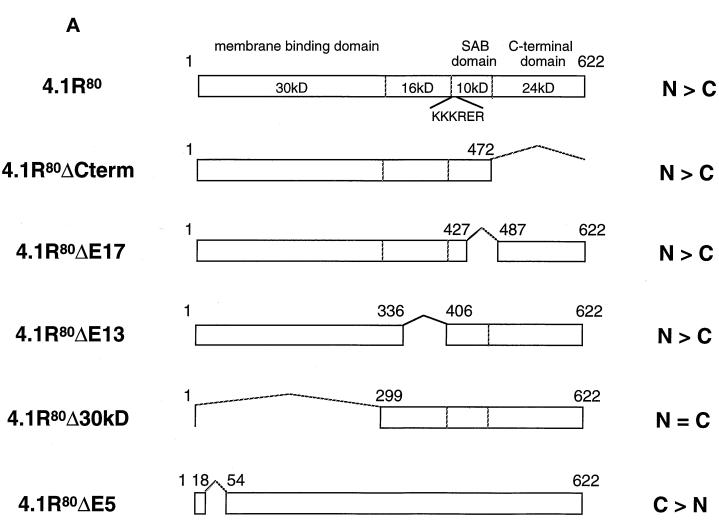
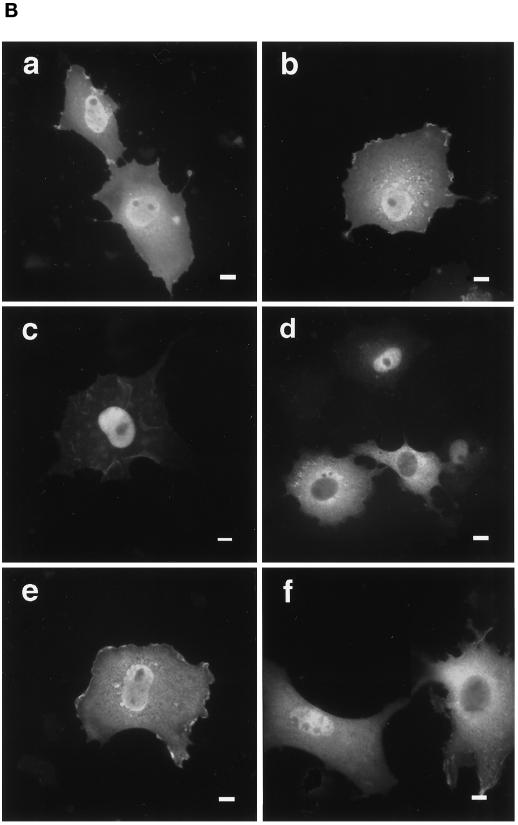
Effects of truncation or point mutation of protein 4.1R domains flanking 4.1R NLS on protein 4.1R nuclear import. (A) Map of protein 4.1R constructs used to investigate nuclear localization. The four chymotryptic fragments of 4.1R80 isoform are shown at the top (30-kDa membrane binding domain, 16-kDa domain, 10-kDa SAB domain [containing the weak core NLS KKKRER], and 24-kDa domain). The protein is 622 amino acids in length. The numbers displayed refer to the amino acids present in each construct. The predominant distribution pattern is displayed for each construct and is based on examination of at least 600 cells per construct: N > C, predominant nuclear staining; C > N, predominant cytoplasmic staining; N = C, equivalent nuclear and cytoplasmic staining. (B) COS-7 cells were transfected with various HA-tagged mutants of 4.1R80. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and then processed for immunofluorescence using an affinity-purified polyclonal antibody against the HA tag as primary antibody and anti-rabbit IgG coupled to FITC as secondary antibody. Cell imaging was performed on samples analyzed by conventional microscopy. Cells were transfected with the following: (a) 4.1R80ΔC-term; (b) 4.1R80ΔE17; (c) 4.1R80ΔE13; (d) 4.1R80ΔE5 (the protein distribution pattern of cells expressing 4.1R80Δ30-kDa domain isoform is similar to that of 4.1R80ΔE5 isoform); (e) 4.1R80mut KH(X)7KR exon5; (f) 4.1R80mut EED exon 5. Bar, 10 μm.