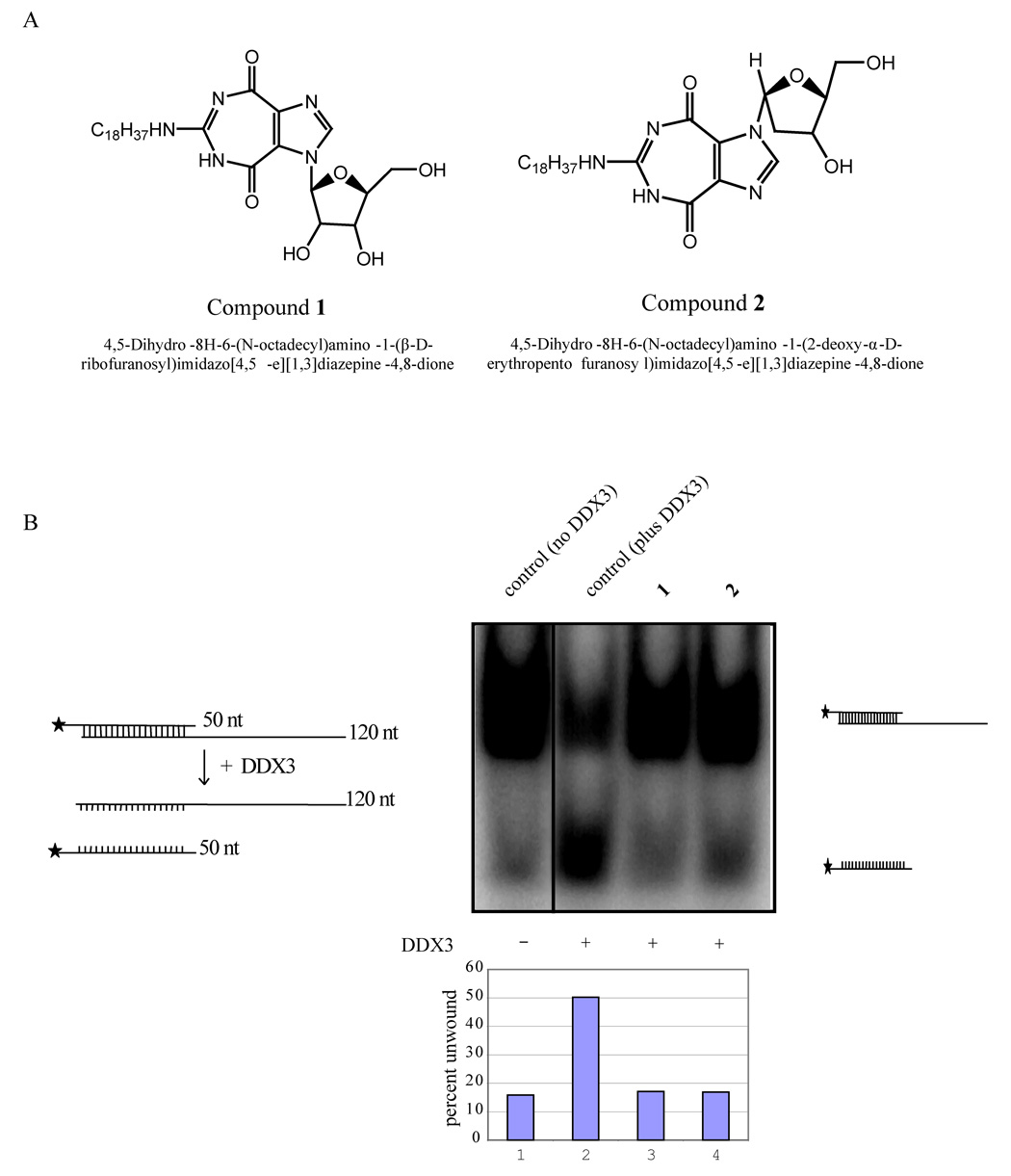
Figure 3.
Inhibition of helicase activity in vitro. A) Structures of 1 and 2. B) Inhibition of helicase activity of DDX3 by 1 and 2. The pBluescript vector was digested to completion with Kpn I and transcribed with T3 polymerase to generate a 120 base long transcript. The vector was also digested with EcoRI and transcribed with T7 polymerase in the presence of (α32p) UTP. The two complementary transcripts (50nt and 120 nt) were purified and hybridized at 95° C for 20 minutes. Note that the 120 nt is unlabeled while the 50 nt strand is 32P-labeled. Autoradiography of the slower migrating band is due to duplex formation between the labeled and unlabeled strands (lane 1). Duplexed RNA was unwound by DDX3 in the presence of ATP resulting in increased release of the labeled strand (50 nt; lane 2) and a reduction in intensity of the duplexed strands. Both 1 and 2 inhibited the ATP dependent RNA unwinding activity of DDX3 as measured by the reduced release of the 50 nt strand (lane 3 and 4). The bar diagram indicates percent duplex unwound by DDX3 in the presence and absence of 1 and 2.