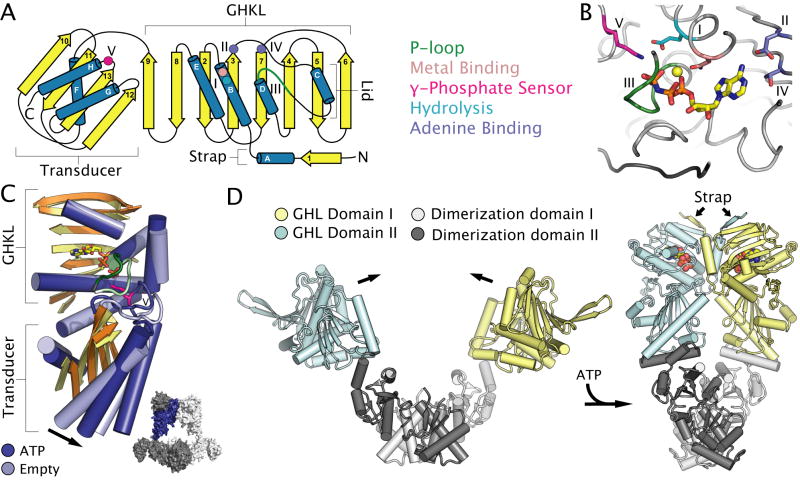
Fig. 3. Nucleotide binding and conformational changes in GHL ATPases.
A. Topology diagram of the two-domain GHL fold. The GHKL and transducer domain are labelled. Conserved motifs, catalytic residues and secondary structural elements are coloured as in Figure 1. Roman numerals define conserved motifs of GHL motors.
B. The active site of A. aeolicus topoisomerase VI (PDB entry 1MX0) (Corbett and Berger, 2003) highlighting the location of catalytic residues common to all GHL ATPases. Active site elements are coloured as in Figure 1.
C. Structural superposition of the GHL fold of ATP-bound (PDB entry 1MX0) (dark blue) and empty (PDB entry 1MU5) (light blue) B-subunits of Sulfolobus shibatae topoisomerase VI (Corbett and Berger, 2003), with most loops removed for clarity. Arrows highlight ATP-dependent conformational changes discussed in the text. Inset: the location of the ATP-bound subunit (shown in dark blue) with respect to the quaternary structure of the topoisomerase VI heterotetramer (with subunits shown in light and dark grey).
D. ATP-dependent conformational changes in a GHL homodimer illustrated by the structures of apo E. coli HtpG (PDB entry 2IOQ) (Shiau et al., 2006) and ATP-bound S. cerevisiae Hsp90 (PDB entry 2CGE) (Ali et al., 2006). ATP is shown as coloured spheres. Subunits of the homodimer are coloured cyan and dark grey, or yellow and light grey, with the cyan and yellow portion of each subunit corresponding to the conserved GHL fold.