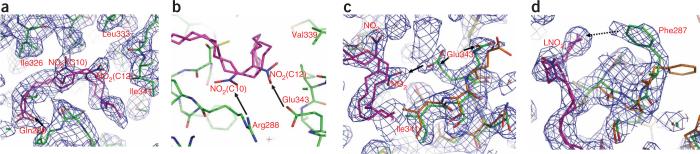
Figure 2.
The structural determinants of the PPARγ LBD and LNO2 complex. (a) 2Fo – Fc electron density map (1.0σ) showing two bound LNO2 isomers and the surrounding PPARγ residues. LNO2 is shown in stick representation with carbon, nitrogen and oxygen atoms depicted in pink, blue and red, respectively. The key residues that determine PPARγ selectivity are noted, and hydrogen bonds are indicated by arrows. (b) Charged interactions of PPARγ residues with specific nitro groups in the LNO2 ligand as determinants of LNO2 selectivity. Hydrogen bonds are indicated by arrows. (c,d) The conformational changes of PPARγ induced by LNO2. Overlays of the PPARγ–LNO2 structure with the PPARγ–rosiglitazone structure, where LNO2-bound PPARγ is in green and rosiglitazone-bound PPARγ is in gold. The conformational shifts of Glu343 toward the nitro group and the shift of Phe287 toward the LNO2 backbone are indicated. The hydrophobic interaction between Phe287 and the LNO2 backbone is shown with a dashed line.