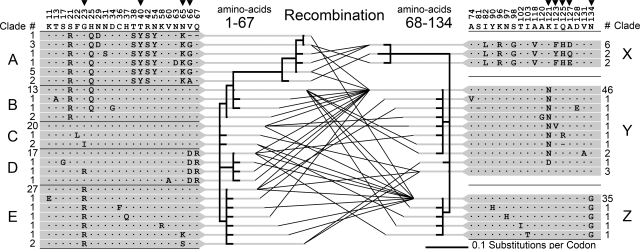
Fig. 4.
Recombination in the F-lectin repeat intron. Amino acid variability among 107 F-lectin repeats of 39 full-length bindin molecules is shown. Numbers above the consensus sequence indicate the amino acid position of variable sites within the repeat. Arrows indicate positively selected sites inferred by PAML. An algorithm that detects recombination finds evidence of a single recombination site between repeat positions 67 and 68. Phylogenetic relationships of the repeat halves on either side of this site are depicted (1–67, 24 unique sequences and 68–134, 18 unique sequences). The lines connecting sequence variants on either side of the site represent the full-length repeats sequenced in this study. Numbers adjacent to each variant indicate the number of times it was sampled. The 5′ and 3′ half-repeats are organized into clades (A–E and X–Z) each composed of a high-frequency allele and its variants. The pairwise combinations of 5′ and 3′ clades generated by recombination are summarized in SI Table 1.