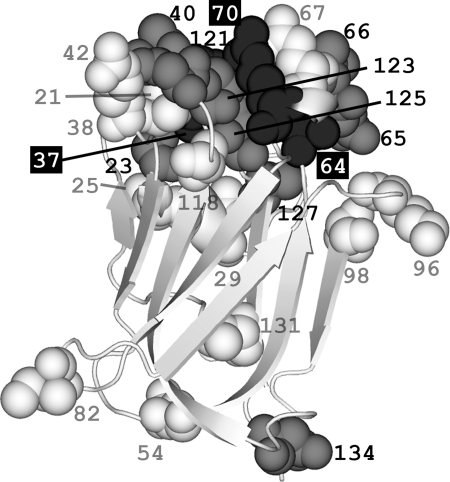
Fig. 6.
Spatial locations of amino acid substitutions in oyster bindin repeats threaded onto the F-lectin crystal structure. Numbers refer to the amino acid position within the repeat. Eight of the nine positively selected sites (estimated dN/dS >1; gray spacefill) are clustered on the sugar binding face of the lectin and surround the 1H and 2R residues of the fucose-recognition motif (black spacefill; Fig. 2). The average distance between these eight positively selected sites is less than that expected by chance (n = 8, P < 0.009, 106 replicates). Polymorphic sites (white spacefill) have amino acid substitutions observed in two or more cDNA sequences. Singleton sites (not depicted) are substitutions observed in only one cDNA sequence. Polymorphic (n = 21, P < 0.783, 106 replicates) and singleton sites (n = 24, P < 0.082, 106 replicates) are not spatially clustered.