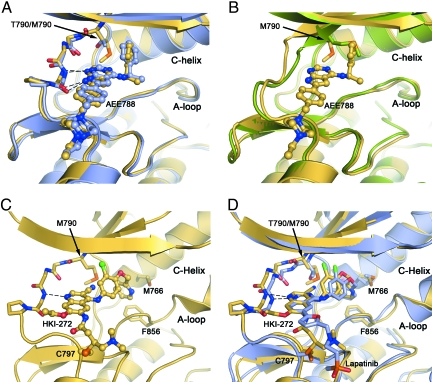
Fig. 2.
Crystal structures of the EGFR T790M mutant show that inhibitors are readily accommodated in the active and inactive conformations of the kinase. (A) Superposition of EGFR T790M/AEE788 complex (yellow) and WT/AEE788 complex [light blue; drawn from PDB ID code 2J6M (8)]. Dashed lines indicate hydrogen bonds to the kinase hinge region that are preserved in both complexes. The location of the T790M mutation is indicated. (B) Superposition of EGFR T790M/AEE788 complex (yellow) and apo-T790M structure (green). Note the alternate side-chain conformation of Met-790 in the presence of the inhibitor. (C) Crystal structure of HKI-272 in complex with the T790M mutant. The kinase adopts an inactive conformation, with the C-helix displaced. A covalent bond is formed between Cys-797 and the crotonamide Michael acceptor of HKI-272. (D) The structure of the T790M mutant in complex with HKI-272 (yellow) is superimposed on the structure of the WT EGFR kinase in complex with Lapatinib [light blue; drawn from PDB ID code 1XKK (32)]. In both structures, the kinase adopts the same inactive conformation and the inhibitors bind in a similar manner, with a single hydrogen bond to the hinge (dashed lines) and with their aniline substituents extending into the enlarged hydrophobic pocket that is characteristic of the inactive conformation.