Fig. 5.
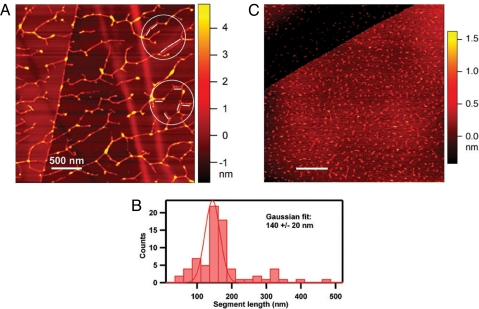
Self-assembled RSH network imaged by AFM. (A) A solution of RSH monomer (9 μg/ml water; see Fig. 3A) was dispersed on a highly ordered pyrolytic graphite (HOPG) surface, and the molecular contours were imaged by AFM. The molecular height of the segments, which ranged from 0.6 nm to 5 nm, was estimated by the colors corresponding to the nanometer ruler on the right. The white bars represent 127-nm lengths and correspond to the calculated length of RSH, assuming a strict polyproline II helical secondary structure (three residues per turn; pitch = 9.4 Å; 404 aa). The segments of the dendritic structure, here aligned with the 127-nm rods (circled), likely correspond to a relatively few overlapping RSH molecules aligned to produce rigid segments slightly longer than the RSH molecule, in keeping with its proposed staggered alignment. (B) Exhaustive analysis of all rigid segments in A yielded the histogram. (C) AFM imaging of (YK)20 (10 μg/ml water), an extensin analog produced through synthetic genes and expressed in BY-2 tobacco cells. (YK)20 contains 20 tandem repeats of the arabinosylated extensin sequence SO4SOSO4YYYK, also in a polyproline II helix. (Scale bars: 500 nm.)