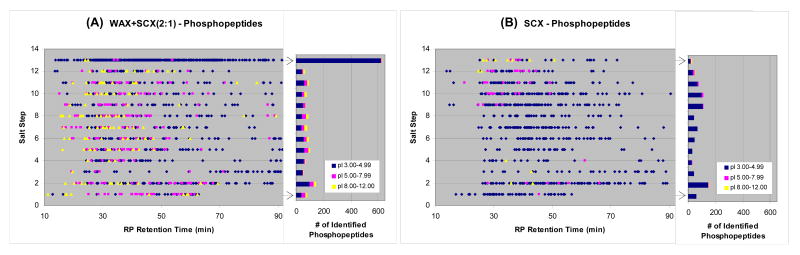
Figure 6. 2D distribution of phosphopeptides identified in (A) WAX+SCX(2:1) and (B) SCX formats using modified 13-step MudPIT.
Phosphopeptides enriched from HeLa cell nuclear extract30 were analyzed in the both ion-exchanger formats (∼50 μg/run). The phosphopeptides were identified from MS/MS spectra of either the precursor peptide or its neutral loss species, and the plots were generated by combining those identifications. The salt pulse for the Step 13 in this analysis was modified as follows: a 2 min salt pulse of 95% buffer D' + 5% buffer B, where buffer D' was water/acetonitrile (95:5, v/v) containing 500 mM ammonium acetate (pH 1.73 by trifluoroacetic acid). Peptide/protein identification criteria were: (1) one or more peptide identification per protein, (2) peptides must be fully tryptic, and (3) estimated false positive rates less than 0.5% at protein level as determined by a decoy reversed-database approach.