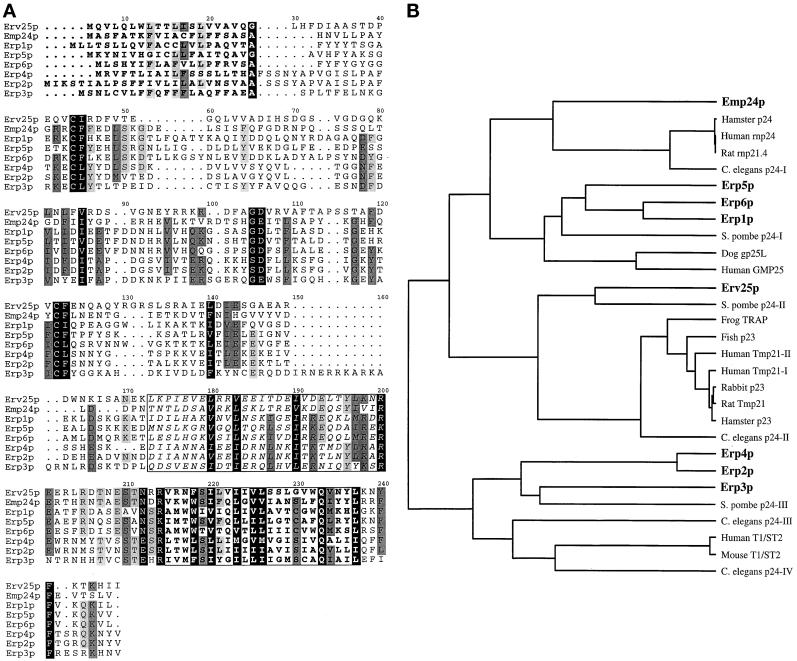
Figure 1.
Yeast p24 proteins and the relationship of currently identified p24 genes. (A) Alignment of the eight p24 members in S. cerevisiae. The predicted N-terminal signal sequences are marked in bold. The putative coiled coil domains are boxed, and in italics and the predicted transmembrane region is boxed and in bold. All yeast p24 proteins, as in other species, contain two conserved cysteine residues within the N-terminal region of the protein that is likely to be in the lumen of a secretory compartment or vesicle. Each yeast p24 protein contains a conserved glutamine residue within the transmembrane domain and a conserved phenylalanine residue (implicated in COPII binding; Dominguez et al. 1998) in the membrane-proximal region of the short cytoplasmic C-terminus. Yeast α family p24 proteins (Erp1p, Erp5p, and Erp6p) contain close matches to the COPI binding motifs KKXX and KXKXX (Cosson and Letourneur, 1994). All the yeast α and β family members (Erp1p, Erp5p, Erp6p, and Emp24p) contain a polar glutamine residue within the transmembrane domain (position 222). This residue in Chop24 has been implicated in ER localization (Fiedler and Rothman, 1997). (B) Relationship between predicted p24 protein sequences currently deposited in public domain databases. The phylogenetic tree shown was generated using the UPMGA clustering method of NEIGHBOR from the PHYLIP package (Felsenstein, 1989), the distance matrix being calculated using PROTDIST.