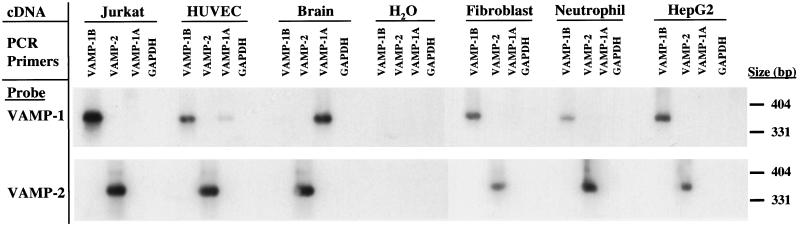
Figure 2.
PCR analysis of the distribution of VAMP-1A, VAMP-1B, and VAMP-2 mRNA. mRNA from Jurkat cells, human endothelial cells (HUVEC), human brain, primary fibroblasts, human neutrophils, and HepG2 cells was prepared and reversed transcribed. The cDNAs and a water control (no cDNA) were then amplified using primers specific for either VAMP-1A, VAMP-1B, VAMP-2, or GAPDH (see MATERIALS AND METHODS). The resulting reactions were separated on agarose gels, blotted to nylon membranes, and then visualized in a Southern blot with probes derived from the complete coding sequences for VAMP-1B (top) or VAMP-2 (bottom). The VAMP-1B probe used here is identical to the VAMP-1A sequence over 339 of 347 bases, and so readily recognizes the VAMP-1A sequence.