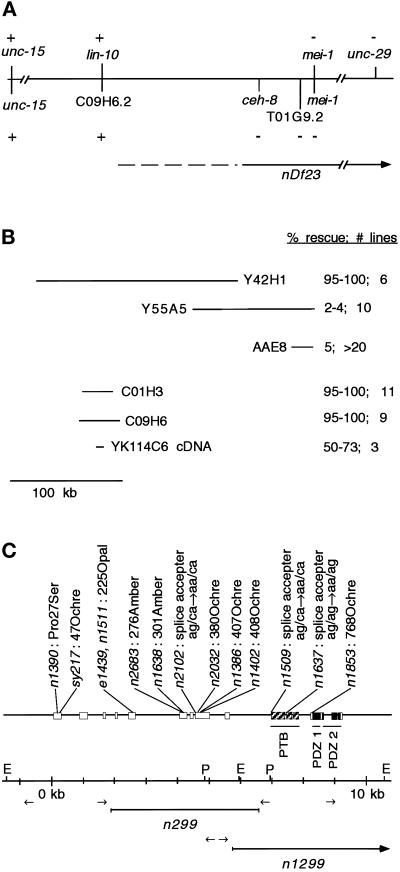
Figure 3.
Molecular identification of lin-10. (A) Genetic and physical maps of the lin-10 region. Genetic and physical markers are shown above and below the line, respectively. (+ and −) Presence and absence of genetic markers (shown by complementation with nDf23) or physical markers (shown using PCR experiments in nDf23 homozygous embryos), respectively. A continuous line, no line, or a dashed line next to nDf23 indicates the region known to be deleted by nDf23, the region known to be unaffected by nDf23, and the region that may or may not be deleted by nDf23, respectively. (B) Genomic and cDNA clones used in transformation rescue experiments. Results of transformation rescue experiments using lin-10(n1390) are shown. Only 0.3% of lin-10(n1390) animals lay eggs.Shown are the percent of transgenic animals that lay eggs and the number of independent transgenic lines. The lin-10 cDNA (YK114C6) was expressed from the vulval-specific lin-31 promoter. Genomic clones Y55A5 and AAE8 also cause a protruding vulval phenotype. Rescue data for cosmid AAE8 was reported by Kim and Horvitz (1990). (C) lin-10 genomic structure was determined by aligning YK114C6 cDNA sequence with the genomic sequence. Exons are indicated by boxes. Regions predicted to encode PTB and PDZ domains are indicated. Positions and molecular lesions associated with lin-10 mutant alleles are indicated. Restriction sites EcoRI (E) and PstI (P) and primer pairs (arrows) used for inverse PCR experiments are indicated.