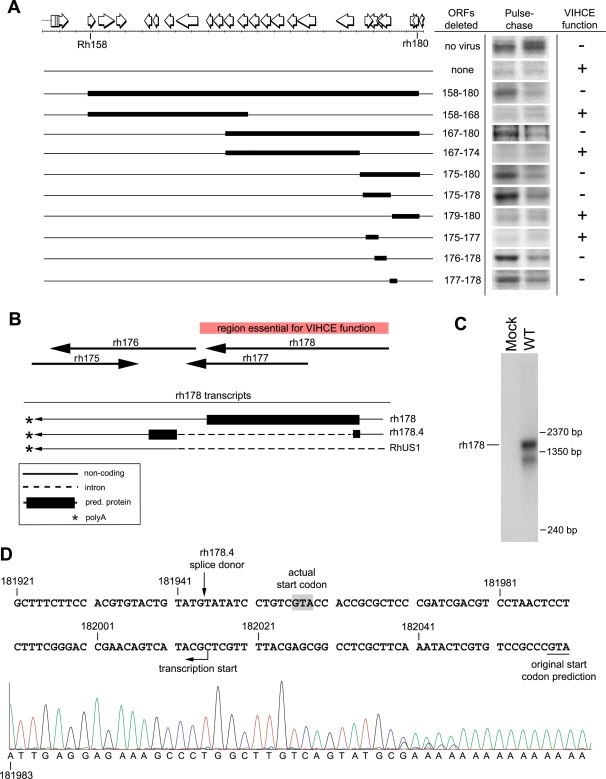
Figure 3. VIHCE is encoded by rh178.
A) Deletional mapping of VIHCE. Predicted open reading frames between Rh158–180 are shown as open white arrows. Solid black rectangles indicate the region of deletion. Pulse-chase labeling for 10 min with the indicated recombinant virus was performed as in Fig. 1C followed by IP with K455. Lack of VIHCE is readily apparent by the initial synthesis of HC (left lane) followed by US2-11-mediated destruction (right lane). B) Predicted ORFs and experimentally confirmed transcripts in the rh178 region. The red rectangle indicates the region essential for VIHCE function as determined by deletions in several independent recombinants. Large black arrows indicate positions of ORFs rh175–178 predicted by [12]. Transcripts confirmed by RACE and cDNA PCR are shown below. C) Northern blot analysis of total RNA isolated from mock or WT RhCMV-infected TRFs at 24 hours post infection. ORF rh178 was used to generate 32P-dCTP labeled DNA probe. D) Complementary sequence of the RhCMV genome from 181921–182060bp. Underlined at 182058bp is the original predicted start codon for rh178 [12]. Transcription actually begins at 182015bp as determined by 5′ RACE (see sequence chromatogram below genomic sequence). Shaded in gray is the first ATG codon of the transcript. Also noted is the splice donor site for rh178.4 which is spliced at 181944bp.