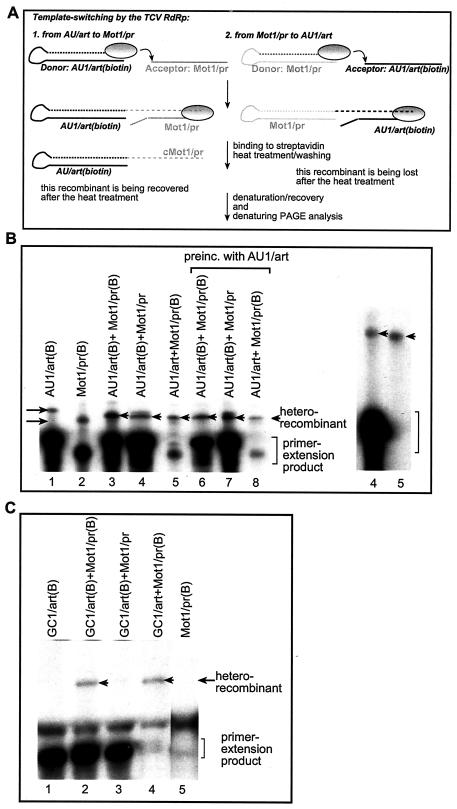
FIG. 4.
Determination of donor versus acceptor use by the TCV RdRp. (A) Schematic representation of the method used for selective purification of recombinant RdRp products. The template switching by the TCV RdRp (shown as an oval circle) is represented by an arrow. The newly synthesized, 32P-labeled RNA sequences are indicated by dotted lines. The two types of heterorecombinants are shown in the left and right panels, respectively. The left panel depicts the scenario when the biotin-labeled RNA template (AU1/art shown as an example) served as the donor RNA during the template-switching event, leading to the formation of a recombinant RNA that carries the label (due to initiation that takes place via 3′ primer extension). The biotin-labeled RdRp products (including the primer extension [data not shown] and the recombinants) were purified using streptavidin as described in Materials and Methods. Note that the heterorecombinants formed between Mot1/pr (donor; unlabeled with biotin) and AU1/art are not labeled with biotin and are lost during purification (as depicted in the right panel). (B) Analysis of the donor template selection by the recombinant TCV RdRp. The RdRp reaction was performed as described in the legend to Fig. 1, except that one of the templates was biotin labeled (indicated by the letter B after the name of the construct) before the reaction. After streptavidin-based purification, the RdRp products were analyzed on denaturing gels. The homorecombinants are depicted with arrows pointing rightward, while the heterorecombinants are marked with arrowheads. Lanes 6 to 8 include samples that were obtained by preincubating the RdRp reaction mixture with the AU1/art template (details are in Materials and Methods). Due to template competition, the primer extension products are lower in the two-template-containing experiments than in single-template-containing experiments. The gel on the right shows a longer run containing samples 4 and 5 to resolve the size differences between the recombinants (marked with arrowheads). (C) Inefficient use of a GC-rich template by the TCV RdRp during template switching. The names of the constructs used are shown above the lanes. See further details in panels A and B. Arrowheads point at the heterorecombinants.