Summary
PhoQ is the transmembrane sensor kinase of the phoPQ two-component system, which detects and responds to divalent cations and antimicrobial peptides and can trigger bacterial virulence. Despite their ubiquity and importance in bacterial signaling, the structure and molecular mechanism of the sensor kinases is not fully understood. Frequently, signals are transmitted from a periplasmic domain in these proteins to the cytoplasmic kinase domains via an extended dimeric interface, and the PhoQ protein would appear to follow this paradigm. However, the truncated periplasmic domain of PhoQ dimerizes poorly when it is excised from the remainder of the protein, so it has been difficult to distinguish the relevant interface in crystal structures of the PhoQ periplasmic domain. Thus, to determine the arrangement of the periplasmic domains of E. coli PhoQ in the physiological homodimer, disulfide scanning mutagenesis was used. Single cysteine substitutions were introduced along the N-terminal helix of the periplasmic region, and the degree of cross-linking in each protein variant was determined by Western blotting and immunodetection. The results were subjected to periodicity analysis to generate a profile that provides information concerning the Cβ distances between corresponding residues at the interface. This profile, together with a rigid-body search procedure, sidechain placement, and energy minimization, was used to build a model of the dimer arrangement. The final model proved to be highly compatible with one of the PhoQ crystal structures, 3BQ8, indicating that 3BQ8 is representative of the physiological arrangement. The model of the periplasmic region is also compatible with a full-length PhoQ protein in which a 4-helix bundle forms in the membrane. The membrane 4-helix bundle has been proposed for other sensor kinases and is thought to play a role in the mechanism of signal transduction; our model supports the idea that signaling through a membrane 4-helix bundle is a widespread mechanism in the transmembrane sensor kinases.
Keywords: PhoQ, disulfide scanning, molecular modeling, two-component signaling, histidine kinase
Introduction
The ability to detect external stimuli is critical for any cell, and especially important for single-cell organisms. In bacteria, most of this sensing is performed by sets of proteins known as two-component systems. There are thousands of different two-component systems spanning all strains of bacteria,1 including about 30 in Escherichia coli alone.2 These systems are involved in detecting and responding to stimuli of all types, including general signals such as osmolarity,3 pH,4 or redox potential,5; 6 and specific signals such as concentration of a particular molecule (citrate,7 or magnesium ions,8 for example).
The two protein components that make up the system typically include a membrane-spanning sensor kinase and a cytoplasmic response regulator.1 The prototypical membrane-spanning sensor kinase, represented by the proteins EnvZ3 and PhoQ,9 consists of three regions. The periplasmic sensor region is responsible for detecting changes in the surrounding medium. Protein sequences and structures from this region share little similarity since they have diverse functions. The transmembrane (TM) region extends the length of the lipid bilayer and serves as an anchor for both the periplasmic and cytoplasmic regions. It typically consists of two membrane-spanning antiparallel helices, located at the N- and C-termini of the periplasmic region. The cytoplasmic region contains a small signaling domain at its N-terminus known as a HAMP domain.10 The HAMP domain, named for its presence in histidine kinases, adenylyl cylcases, methyl-accepting proteins, and phosphatases, is widespread in a variety of proteins and plays a role in transducing the signal from the periplasm into catalytic activity. The remainder of the cytoplasmic region is a well-conserved histidine kinase domain that interacts with the response regulator protein. Upon activation, the kinase first autophosphorylates a conserved histidine and subsequently transfers the phosphoryl group to an aspartate on the response regulator. The phosphorylated regulator protein then typically binds to DNA and promotes transcription of one or more genes. Many variations on this prototypical sensor kinase architecture are known, including missing or additional domains and alternative sensing mechanisms.1
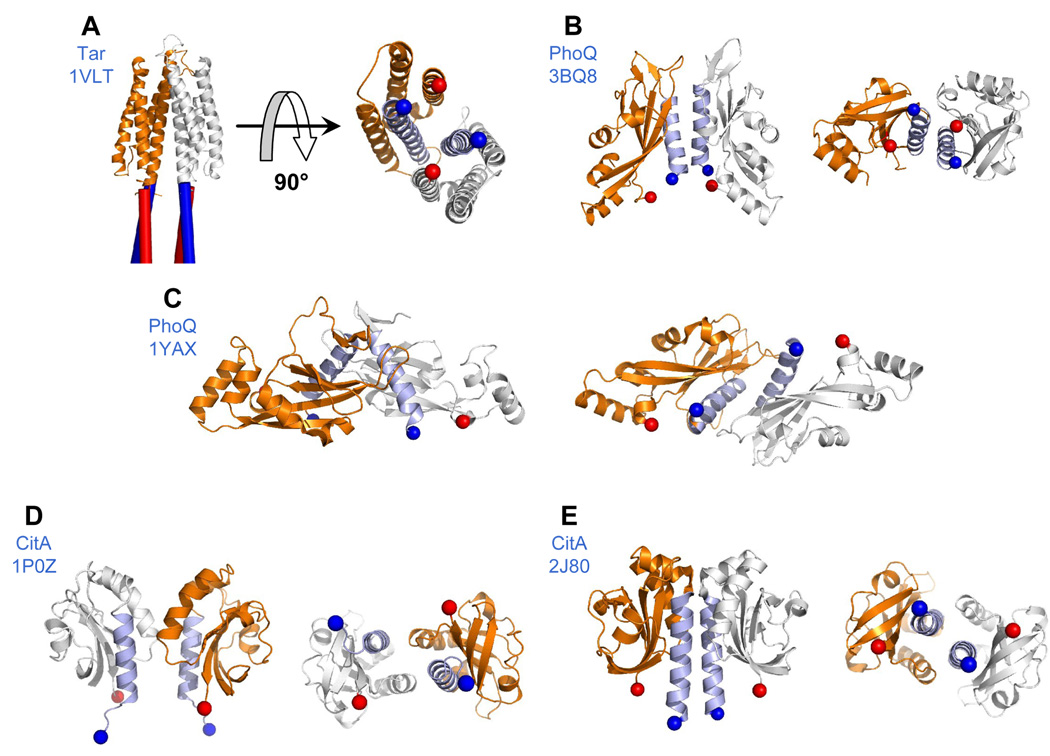
Elucidation of the structure and precise molecular mechanism of signaling in the sensor kinase molecule has been the focus of a number of studies.11; 12 Tar, the aspartate receptor that mediates chemotaxis in E. coli and Salmonella enterica serovar typhimurium, is the best studied sensor kinase-like protein. Tar has similar architecture to the sensor kinases, though the cytoplasmic region lacks a kinase domain (and instead forms a complex with a cytoplasmic kinase). The full-length Tar protein is a stable homodimer in vivo,13 and the isolated periplasmic domain forms dimers in solution.14 The crystal structure of the Tar periplasmic domain shows a dimer of 4-helix bundles15 (Figure 1a). Helices α1 and α4 from each subunit are located in the interior of the dimer, with their N-termini 17 Å apart and their C-termini 18 Å apart. This suggests that the transmembrane (TM) helices TM1 (which leads into helix α1 and is colored blue in Figure 1a) and TM2 (which extends from helix α4 and is colored red in Figure 1a) from each subunit form a 4-helix bundle in the membrane, and extensive disulfide cross-linking studies on the TM domains have provided strong support for this hypothesis.11 This transmembrane 4-helix bundle (henceforth ‘TM bundle’) model requires that the corresponding N- and C-termini of the periplasmic domain are separated by about 15–20 Å in order for the transmembrane helices to approach each other closely enough to associate. The model has led to a proposed mechanism in which substrate binding causes a conformational change in the periplasmic domain which is propagated along a continuous helix through the TM bundle and into the HAMP domain, thereby altering the signaling state.11 Proposals for this conformational change include a piston shift,11 a helix rotation,16 or unwinding of a coiled coil.17
Figure 1.
Dimer configurations of sensor kinase periplasmic domains. PDB IDs appears in parentheses. (A) Tar (1VLT) - cylinders depict the proposed 4-helix TM bundle that extends from the periplasmic domain; (B) PhoQ (3BQ8); (C) PhoQ (1YAX); (D) CitA (1P0Z); (E) and CitA (2J80). Each dimer configuration is depicted in two panels. The left panel shows the protein from the “side” view, sitting on top of the membrane, as it would be seen if viewed from the plane of the membrane. The right panel is rotated 90 degrees and depicts the “bottom” view of the protein, as it would be seen if looking through the membrane from below. N-terminal helices are colored light blue. Blue and red spheres indicate the N and C-termini respectively. N-terminal distances are: (A) 17 Å (B) 18 Å (C) 27 Å (D) 30 Å and (E) 15 Å. C-terminal distances are: (A) 18 Å (B) 18 Å (C) 50 Å (D) 29 Å and (E) 30 Å.
While great advances have been made toward understanding the signaling mechanism of Tar, it is unclear whether other sensor kinases similarly function via signal propagation down a helix and through a TM bundle. A number of structural studies have been carried out on the sensor kinases EnvZ, LuxQ, PhoQ, and CitA. The EnvZ protein was shown to be a dimer in vivo, and the isolated periplasmic domain is dimeric in solution.18 Cross-linking studies demonstrated that the N-terminal helices of the periplasmic region are closely associated in the full-length protein19, which would position them appropriately to extend into a TM bundle. The LuxQ, PhoQ and CitA proteins all contain PAS20 or PAS-like21 domains, in which dimerization via an N-terminal α-helix is a fairly common theme.22; 23 The periplasmic portion of the LuxQ protein was shown to be dimeric in solution in the presence of the LuxP protein and the AI-2 inducer, and a crystal structure of the complex showed close association of the N-terminal helices of LuxQ.24 Studies on PhoQ and CitA, though, are less clear. Both PhoQ and CitA feature cytosolic HAMP10 and DHp domains,25 which are indicative of dimerization. In both cases, though, in contrast to Tar and EnvZ, the isolated periplasmic domains do not dimerize strongly in solution.9; 26; 27 Further complications arise from analysis of the crystal structures of these domains.
Two crystal structures of the PhoQ periplasmic domain have been solved, which have led to two opposing models for the dimerization interface of this protein. A crystal structure of the E. coli periplasmic region (3BQ8, solved by Cheung et al.28) features a dimer in the asymmetric unit, in which the N-terminal helices are closely associated. The N- and C- termini are all in close proximity, and the structure is compatible with the TM bundle model (Figure 1b). The 3BQ8 structure also features an inter-subunit salt bridge between residues Arg50 and Asp179 whose physiological relevance is supported by genetic evidence.28 In contrast, a crystal structure of the S. enterica protein (1YAX, solved by Cho et al.9) features 4 monomers in the asymmetric unit, providing a number of potential interfaces when interactions of monomers within different unit cells are considered. One dimer in these structures has been proposed to be the physiological arrangement; this dimeric arrangement is attractive and has been widely adopted as a working model,1; 21; 29 because it helps explain how PhoQ might respond to divalent cations. However, the dimeric arrangement has a very large distance between the corresponding termini (27 and 50 Å), which clearly is not compatible with the TM bundle model (Figure 1c). The S. enterica and E.coli proteins have a very high degree of sequence identity (85%), so it is unlikely that these crystal structures reflect actual differences in the quaternary structures of the full-length proteins. Thus, it was particularly critical to assess which of the two working models is correct.
A similar situation also holds for CitA. Two sets of crystal structures of the Klebsiella pneumoniae CitA periplasmic domain have been solved. The first structure featured an asymmetric unit containing 10 different monomers, and a number of possible dimer arrangements.27 One of these, consisting of chains G and J, features the N- and C-termini in close proximity and is compatible with the TM-bundle model (Figure 1d). However, the more recent set of structures30 is also compatible with the TM-bundle model but features very different subunit contacts from the original structure (Figure 1e). This casts some doubt on the relevance, with regard to physiological arrangement, of the crystal structures of these periplasmic domains that do not dimerize in solution.
To resolve the conflicting data, we decided to determine the arrangement of the periplasmic domain of PhoQ within the full-length protein. PhoQ is a particularly interesting sensor kinase. It is a component of the phoPQ system, which also consists of the PhoP response regulator.31; 32 The primary function of phoPQ is to sense divalent cations (Mg2+ and Ca2+) and activate transcription of a set of genes upon deprivation, such as mgtA, which encodes a magnesium transporter protein.33 Furthermore, in some species of bacteria, such as S. enterica serovar typhimurium, phoPQ is also responsible for orchestrating the expression of genes required for virulence.31; 32 S. enterica PhoQ is activated by cationic antimicrobial peptides of the innate immune system, which have been proposed to bind directly to the periplasmic sensor domain and activate the kinase.34 Activation initiates host cell invasion, and also triggers resistance mechanisms such as cell-surface modification. PhoQ is thus a potential antimicrobial target, as inhibition of signaling could prevent bacterial virulence. Understanding PhoQ signaling on a molecular level could also aid in efforts to design analogs to the natural antimicrobial peptides that retain the antimicrobial properties but do not trigger virulence. Both PhoQ inhibitors and antimicrobial peptide mimics could be important complements to currently available antibiotics in combating the proliferation of resistant strains, such as methicillin-resistant Staphylococcus aureus (MRSA), that has emerged as a major global health threat.35
To elucidate the arrangement of the PhoQ periplasmic region of the full-length protein, we used a combination of disulfide scanning and molecular modeling. This process generated a model of the PhoQ periplasmic region, arranged as a homodimer, which is representative of the native structure in vivo. The dimer that emerges from these studies is structurally distinct from the model first proposed by Cho et al,9 but nevertheless is consistent with their original proposition concerning the interaction of membrane-associated Mg2+ ions and an acidic patch in PhoQ. Thus, it leads to a refinement of our understanding of the mechanism of signaling in response to Mg2+.
Disulfide scanning
A set of single-cysteine variants was constructed spanning the N-terminal helix (residues 47–62) of the periplasmic domain. Since there are no cysteines in the native PhoQ periplasmic domain, each of these variants would then have a total of two cysteine residues in the periplasmic region of the putative PhoQ homodimer. Cysteine residues in the periplasm, which is an oxidative environment, will form disulfide cross-links at a frequency roughly correlating to the distance between them.36 Thus, determining the extent of cross-linking at a series of residues provides a map of the relative spatial arrangement of the PhoQ subunits.
To facilitate analysis of the extent of cross-linking, the E. coli phoQ gene was modified with a C-terminal 6-histidine tag to allow immunodetection, and placed under a trc promoter on the plasmid pTrc99a37 to facilitate genetic manipulation. Sixteen sequential single-cysteine phoQ mutants covering positions 47 through 62 were constructed by site-directed mutagenesis. These positions cover most of the predicted helix that emerges from the membrane, which is believed to occur near residue 40 (±5 residues).26 PhoQ-His6 and the 16 cysteine variants were expressed in E. coli strain TIM206 (obtained from Tim Miyashiro and Mark Goulian), which has the native phoQ gene deleted, and has a lacZ reporter for phoQ activation consisting of lacZ under the control of the phospho-PhoP-induced mgtA promoter. The strains constructed were assayed for PhoQ activity using the standard lacZ assay38 to ensure that the protein variants remain functional, and then assessed for cysteine cross-linking.
Native E. coli PhoQ expressed from the pTrc plasmid shows induction in low Mg2+ media of 3 to 3.5-fold relative to high Mg2+ media. The His-tagged version of PhoQ shows a slight decrease, with induction of around 2.5-fold. In the absence of functional PhoQ protein, negligible β-galactosidase activity is detected. All 16 mutants were functional with varying levels of β-galactosidase activity (Figure S1), indicating that all of the PhoQ variants retain the same overall structure as the wild-type protein. Variation in activity is commonly seen in disulfide scanning39, and can potentially provide further structural and mechanistic information which we will explore in future studies.
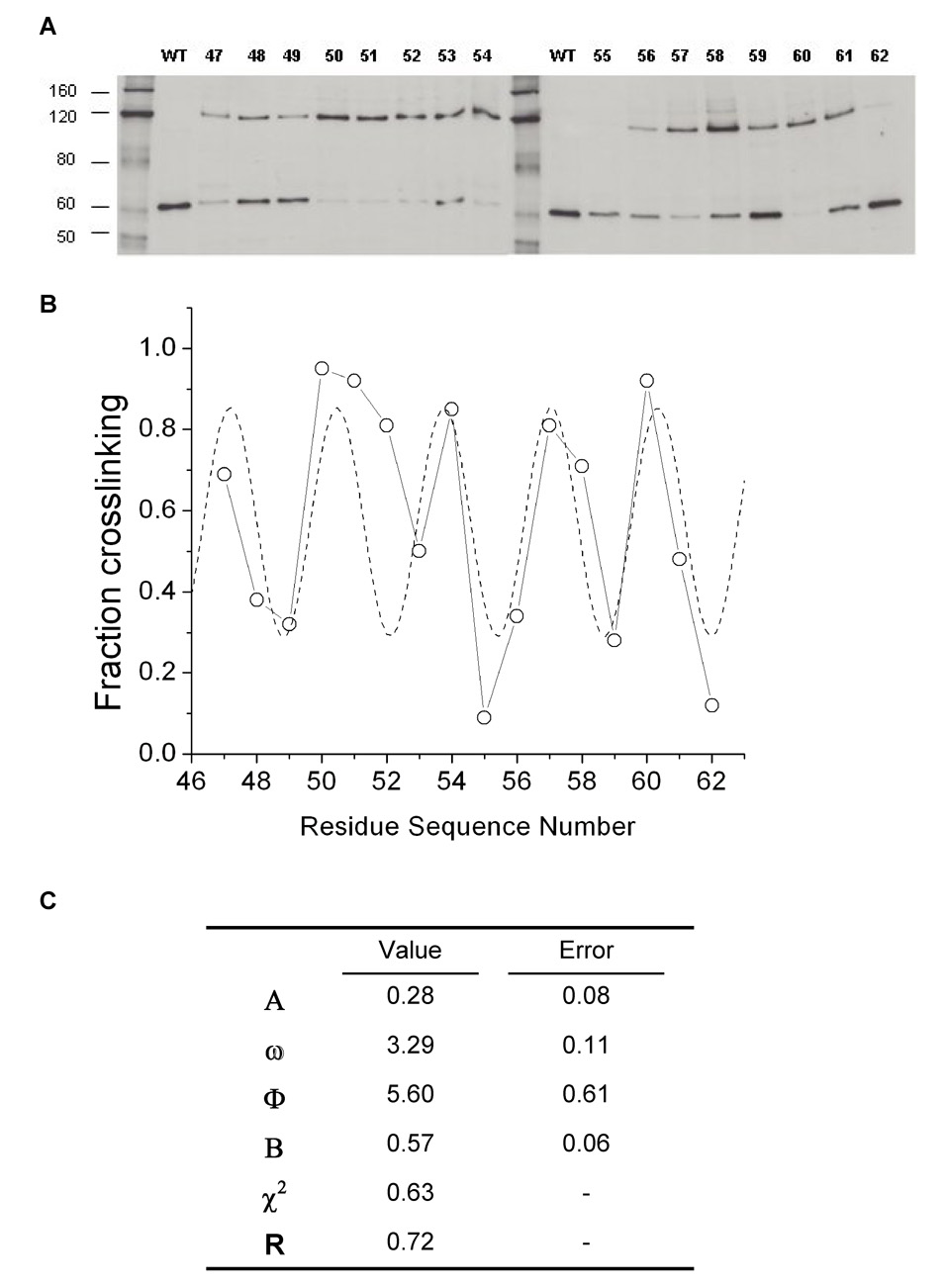
The degree of cross-linking in each variant was determined by immunodetection with anti-His tag antibodies (Figure 2a). Cells expressing each protein were grown in LB media and the cell envelopes, which are enriched in PhoQ protein, were isolated. N-ethyl maleimide (NEM) was used to alkylate all free cysteines to ensure that no further cross-linking occurred after the cells were harvested. The envelope preparations were subjected to denaturing SDS-PAGE and transferred to nitrocellulose membranes by Western blotting. The membranes were probed with the Penta-His primary antibody (Qiagen), an HRP-linked secondary antibody, and ECL detection reagents. The amount of protein corresponding to monomer (~55 kD) and to cross-linked dimer (~110 kD) was quantified for each mutant using the Scion Image imaging software (Figure 2b). The preparations shown were obtained from cells that were grown to stationary phase. Similar samples from cells harvested at mid-log phase yielded qualitatively similar monomer-dimer ratios. However, we found that the expression of PhoQ in our system was higher in saturated cells and thus yielded much clearer and more easily quantifiable blots.
Figure 2.
Fraction of cross-linked residues. (A) Immunoblots of SDS-PAGE gels displaying envelope preparations of cells expressing His-tagged PhoQ variants. Cell envelopes were isolated using a protocol modified from Morona and Reeves.46 After the final spin to harvest the envelopes, pellets were resuspended in 8 M urea. Gel samples were prepared in 8M urea and NuPage LDS loading buffer (Invitrogen), and run on 7% Tris-Acetate SDS-PAGE gels (Invitrogen). Protein was transferred to nitrocellulose membranes, blocked with 3% BSA, and then incubated with Penta-His anti-His tag antibody (Qiagen) diluted 1:2000 for 16 hrs. at 4°C. After washing, blots were incubated with HRP-linked anti-mouse IgG (GE Bioscience) diluted 1:5000 and developed with ECL reagent (GE Bioscience). All 16 mutants are shown, as well as wild-type PhoQ. The Benchmark His-tag protein ladder (Invitrogen) was used as a molecular weight standard, with the approximate molecular weights indicated. Bands corresponding to PhoQ monomer (~55 kDa) and cross-linked dimer (~110 kDa) are shown. (B) Best fit curve (---) obtained by fitting the percentage of cross-linking between residues to equation [1]. Percentages were obtained by quantitation of the monomer and dimer bands on a PC computer using the Scion Image software, a PC version of the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). (C) Parameters from fit of cross-linking data to equation [1]. ω is the repeat, related to secondary structure and the helix-crossing angle. Φ is the phase, indicating which face of the helix is directed towards the helix-helix interface. B is the mean cross-linking efficiency. A is the variation in efficiency between residues on opposite sides of the helix.
Many of the cysteine variants cross-linked efficiently without addition of an oxidation catalyst. This is not always seen in disulfide scanning studies, even in the oxidative environment of the periplasm, and is thought to occur only at regions of tight and stable association of subunits.40 Quantitation of the extent of dimerization at each of the analyzed positions showed a clear periodicity, with peaks occurring at positions 47, 50–51, 54, 57, and 60, which lie along one face of an α-helix. This finding strongly suggests that the scanned region is helical, as in the crystal structures of PhoQ. Furthermore, the mean cross-linking efficiency is around 0.5, ranging from almost 100% efficiency at the peaks to almost no cross-linking at the lowest points. This pattern is very similar to that expected for a tightly packed parallel helix-helix interface, as has previously been observed in other structures.41 Finally, it is interesting to note that there is no net overall trend towards greater cross-linking as one progresses from one end of the scanned region to the other (with the exception of the pronounced α-helical periodicity). This observation suggests that the helices are well packed throughout the length of the interface, which is in good agreement with the crystallographic dimer shown in Fig. 1b. However, it is less concordant with the expectations from the crystallographic dimer in Fig. 1c, in which the N-terminal helices are in van der Waals contact at their C-terminal ends but separated by 30 Å at the N-terminal ends.
Periodicity analysis
While the qualitative results of cross-linking are sufficient to support one crystallographic dimer over another, it is important to ask whether this model is uniquely able to explain the cross-linking data, or whether other unanticipated models might also be able to explain the results. Thus, using the crystallographic monomer of the PhoQ periplasmic domain, we built an independent model of the dimer. We first analyzed the periodicity of the cysteine-scanning mutagenesis data as a function of residue position. We then used this periodicity together with a rigid-body sampling protocol to reorient the N-terminal helices of PhoQ. Finally, these models were subjected to repacking and energy minimization, yielding a final ensemble of models that was most consistent with the experimental results.
Periodicity analysis of the cross-linking efficiency can provide important information concerning the geometry of helix-packing interfaces.42 In a parallel helical dimer, the probability of two sidechains being close together should vary sinusoidally with the residue position along the chain (n), giving rise to periodic changes in cross-linking efficiency according to equation [1]
| (1) |
Nonlinear least-squares fitting of these parameters to the experimental crosslinking data provides: ω, the repeat, which relates to the secondary structure and the helix-crossing angle; Φ, the phase, which relates to which face of the helix is directed towards the helix-helix interface; B, the mean cross-linking efficiency, relating to the mean interhelical distance; and A, the variation in efficiency between residues on opposite sides of the helix (Figure 2c).
The experimental data are very well described by eqn. [1] with a correlation coefficient of −0.7 (p < 0.01 using a two-tailed test). As in any scanning mutagenesis experiment, the fit is not perfect because of experimental uncertainties and the heterogeneous nature of the interface. In addition to the distance between interacting groups, the extent of cross-linking will reflect differences in surrounding steric bulk, the pKa of the Cys thiols, flexibility, and the directions in which the sidechains project. Nevertheless, the fitted repeat period of 3.3 ±0.1 residues was found to be significantly less than that of the α-helix (3.6 residues). A value of less than 3.6 residues is characteristic of a left-handed helical crossing angle as in classical helical bundles and coiled α-helix dimers.43
We next searched for dimeric models whose Cβ-Cβ distances would best correlate with the experimental observations while maintaining reasonable energetic interactions. The interhelical Cβ-Cβ distances, when considered as a function of position are very sensitive to dimer orientation (Figure S2) and should match the same trends as in the experimental data. Thus, minima in the Cβ-distance profiles will correlate with regions of maximal cross-linking while maxima will correspond to positions of minimal cross-linking. To generate models that best correlate with the periodicity analysis we next: 1) performed rigid body sampling to generate an ensemble of dimers; 2) filtered the ensembles to assure reasonable geometries; 3) minimized the structures; and 4) clustered the resulting structures to define ensembles of models that agree with the experimental data.
Model construction
We performed a rigid-body search using chain A from the Cho et al. (1YAX) structure. All monomers in the Cho and in the Cheung et al. (3BQ8) crystal structures are superimposable to a Cα root-mean-square distance (CRMSD) of about 1.0 Å or less (excluding residues 72–84 which make up a mobile loop) and thus are essentially interchangeable for this purpose. The N-terminal helix, which is slightly curved in the Cho crystal structure, was replaced by an ideal α-helix to avoid biasing the search. Finally, the helix was stripped of all sidechains. The rigid-body search resulted in multiple dimer arrangements that were evaluated energetically.
The rigid-body search (Figure S3) begins with the alignment of the N-terminal helix of the monomer onto the global Z-axis. This alignment, along with all rotations and translations, were applied to the entire monomer as a rigid body. After centering the N-terminal helix, the entire monomer was translated along the global Z-axis (Ztrans) followed by translation along the global X-axis (Xtrans). After translation, the monomer was rotated about the global X-axis (Xrot). Finally, the monomer was allowed to rotate about the central axis of the N-terminal helix (Axialrot) to vary the phase of the helix. Due to the C2 symmetry, these 4 degrees of freedom are sufficient to sample all regions of space. The parameters were systematically varied as follows: (a) Ztrans was varied between −10 Å and 10 Å using a step size of 1.0 Å; (b) Xtrans was varied between 3.0 Å and 7.0 Å using a step of 1.0 Å; (c) Xrot was varied between −50 and 50° using a step size of 1.0°; and (d) Axialrot was varied between −180° and 180° using a step size of 5.0°. The symmetry mate was then generated by applying a 180° rotation about the global Z axis. The search generated 774, 165 dimer configurations that were screened using two geometric filters.
The first filter screened for steric clashes between the two monomers. If two heavy atoms from separate monomers were within a distance of less than 4.0 Å, the model was discarded. The second filter used the r value between the best fit curve derived from the percentage of cross-linked residues (see Figure 2b) and the corresponding Cβ distances from the N-terminal helices of the models. Models were discarded if their correlation coefficients were greater than −0.9. After application of the geometric filters, 848 models remained.
To reduce the number of models that needed to be considered for energetic evaluation, they were first clustered according to the CRMSD over the N-terminal helices. Our method of clustering is identical to single-linkage clustering with the following exception: After a cluster has been populated so that no other models can be placed into the cluster below some CRMSD threshold (in this case 1.0 Å), the models in the cluster are removed from the data set and clustering starts over again. Application of this type of clustering to the 848 models resulted in 28 clusters. The member from each cluster with the smallest sum of CRMSD values to all other members in the cluster was selected as the centroid.
The next step in generating a model of the dimer was repacking the side chains of the N-terminal helices. Side chains were added to the N-terminal helices of all 28 centroids using SCAP,44 a pseudo-Monte Carlo procedure with a simplified heuristic scoring function. Since the backbone geometry was kept rigid, side chain addition occasionally resulted in steric ‘bumps’ that were relieved using energy minimization using 1000 steps of Powell energy minimization with the CHARMM22 molecular mechanics force field implemented in the XPLOR-NIH package.45 We used a distance dependent dielectric (DDD) set to a value of 4r to mimic the effect of the solvent. A dihedral restraining function with an energy constant of 10 kcal/mol-rad2 with ±2 degree restraint was used during the energy minimization to prevent large movements in the native structure.
After side chain addition and energy minimization, the stability of each dimer was assessed using the rigid-body minimization (RBM) procedure in the XPLOR-NIH package. RBM attempts to minimize the energy of the system using 3 rotational (Eulerian angles) and 3 translational degrees of freedom. Since we were concerned with dimer stability, only the non-bonded energy terms from the CHARMM22 force field were used. If the dimer interface was unstable due to poor packing, the RBM procedure resulted in large displacements from the starting pose. Models were discarded if the RBM resulted in a displacement larger than 1 Å from the starting pose (computed over Cα atoms for residues 45–62). Eleven of the centroids were discarded due to large RBM displacements, leaving 17 models remaining. This ensemble of models is structurally similar to 3BQ8, ranging in CRMSD from 1.6–3.2 Å over the N-terminal helices. Thus it is more representative of the dynamic nature of the PhoQ interface than any single model. For the purposes of comparison with 3BQ8 and 1YAX, we chose centroid 1 - model 734 (Figure 3) - as the final model since it had significantly lower energy than the rest (see Table SI).
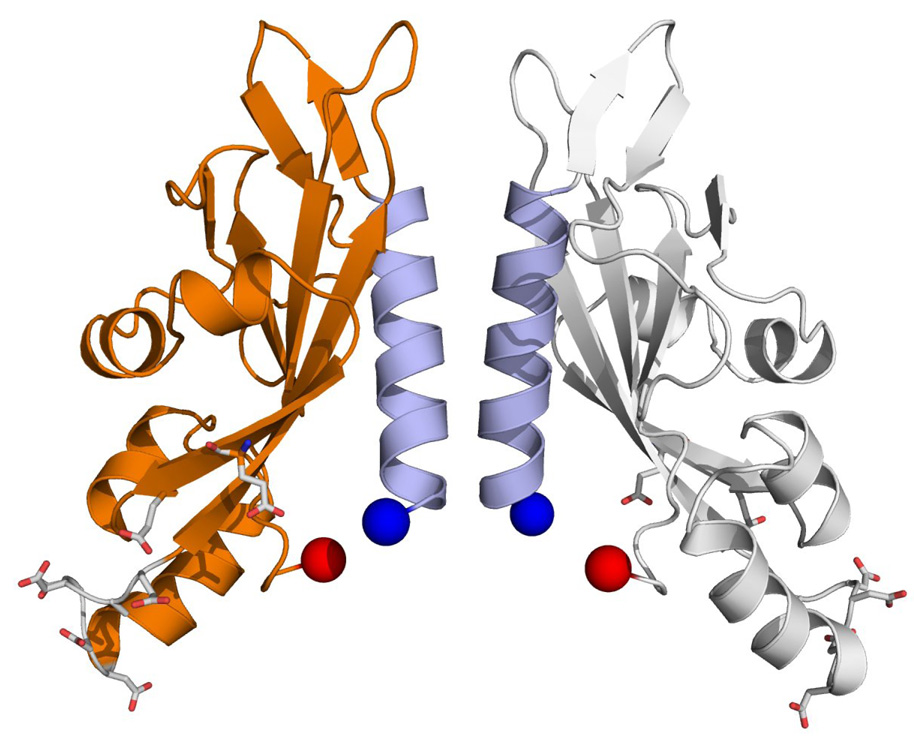
Figure 3.
Best scoring model of the PhoQ periplasmic domain. Model 734, after rigid-body minimization with XPLOR-NIH, is shown. The N-terminal ends are 16 Å apart. The C-terminal ends are 22 Å apart. N-terminal helices are colored light blue. Negatively charged residues implicated in forming interactions with the membrane are shown in stick representation.
The final model is remarkably similar to the Cheung crystal structure. The N-terminal helices of model 734 and those of the Cheung structure superimpose with a CRMSD of 1.9 Å. The N-terminal ends of the model are 16 Å apart measured between the Cα atoms of residue 45 and the C-terminal ends are 22 Å apart measured at residue 186. Similar measurements for the Cheung et al. crystal structure indicate that both N and C-termini are 18 Å apart. The helical crossing angle, relative to the dimer axis, is approximately 18°; the corresponding angle for the Cheung structure is about 21° (Figure S4). Thus, both the Cheung structure and model 734 would be poised for entry into the membrane at an angle of roughly 20° relative to the bilayer normal. This would potentially allow residues from both N-terminal and C-terminal helices to interact inside the membrane. The model differs from the Cheung structure to a greater degree on the periphery of the protein. This can be attributed partly to the asymmetry between the monomers of the Cheung structure.
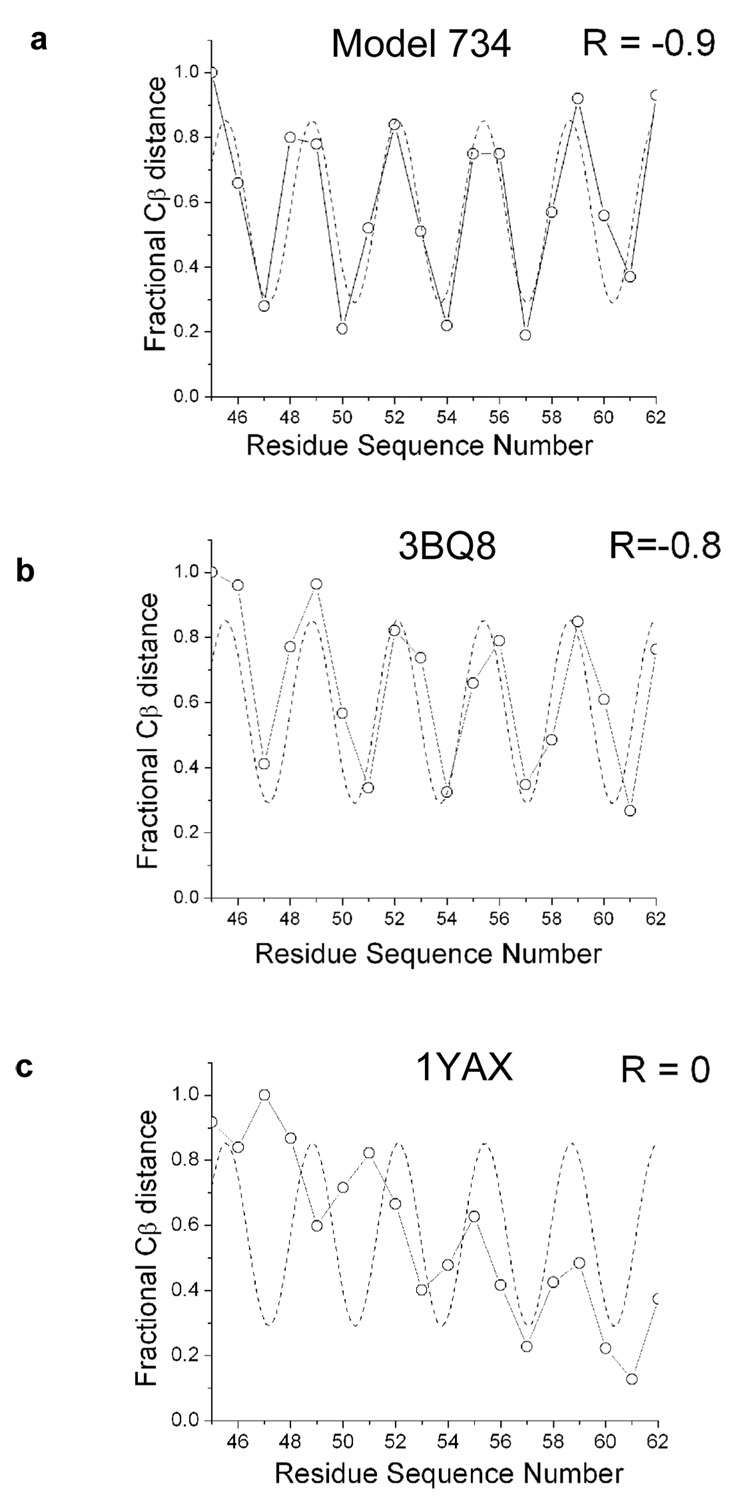
Dimer configurations for both model 734 and for the Cheung structure are both in excellent agreement with the theoretical cross-linking curve (Figure 4a and b). The r values computed between the Cβ distances and the curve for both model 734 and for the Cheung structure show a strong anti-correlation. This is as expected, since atoms that are closest in space should have the strongest cross-linking signal. In contrast, the Cβ distances from the Cho structure yield a poor fit to the cross-linking data (Figure 4c). The poor fit from the Cho structure comes from the large Ztrans and Xrot values (approximated using the best fit helices with a CRMSD of 0.7 Å). The Ztrans value is about −10 Å (as opposed to 2 Å for the Cheung structure and model 734), placing the point of closest approach toward the C-terminal end of the N-terminal helices. The crossing angle is nearly 90 degrees, resulting in large Cβ distances at the N-terminal end of the helix and smaller distances at the C-terminal end.
Figure 4.
Periodicity plots. Periodicity versus Cβ distance is shown for (a) Model 734 (b) 3BQ8 and (c) 1YAX. The best fit curve (---) was obtained by fitting the percentage of cross-linking between residues to equation [1] and then shifting the phase by π to transform the data from being anti-correlated to correlated. Fractional values were obtained by dividing all the Cβ distances for a set by the largest distance in the set. The distance between Cα atoms was used for glycine 54. Correlation coefficients (R) between distances and the theoretical cross-linking curves are shown.
It is interesting to note that the N-terminal helices of model 734 and of the Cheung structure approach each other most closely at the Gly54 residue located approximately at the center of the helix. The mechanism of signal transduction in PhoQ likely involves re-orientation of the two subunits relative to one another. Having the closest contact of the subunits at a glycine residue, then, may play an important role in this movement; glycine lacks a sidechain, and so the helix interaction is not secured by a zipper or knob-into-hole type interaction. This may be important in allowing sliding or pivoting of helices in the course of PhoQ signaling.
Conclusions
We have constructed a model of the periplasmic region of PhoQ under physiological conditions using restraints from our disulfide scanning experiments. The model conforms remarkably well to the Cheung et al. PhoQ crystal structure 3BQ8, indicating that this crystal structure is indeed representative of the quaternary structure of the full-length protein under physiological conditions. Both the Cheung structure and our model are consistent with close positioning of both the N-terminal and C-terminal TM helices, as in the transmembrane 4-helix bundle model described for the Tar protein. Specifically, the Cheung structure and our cross-linking experiments both place the C-terminal ends of TM1 and the N-terminal ends of TM2 in close proximity. Finally, the presence of a HAMP domain that is continuous with TM2 places another restraint on the structure of the TM four-helix bundle. Together, these data strongly support the idea that the TM bundle model is broadly applicable to many two-component sensor kinases.
Finally, it is important to consider the implications of our model with respect to the mechanism of activation of PhoQ proposed by Cho et al. Based on the dimer observed in 1YAX, these authors predicted that Mg2+ and other cations could hold the protein in an inactive state by bridging between the acidic lipid headgroups in a bacterial periplasmic membrane and a carboxylate-rich patch in PhoQ. Our model retains the proximity of these patches to the membrane, although the patches are much closer to one another and approach the membrane at a somewhat different angle. Thus, it is important to consider the possibility that the differences between the two crystallographic dimers reflect different activation states of PhoQ. However, the 1YAX dimer is clearly not consistent with a transmembrane dimer and would require dissociation of the HAMP and kinase domains. While this dissociation is unlikely, it remains possible that a displacement that is less extreme than that implied by the 1YAX dimer underlies signal transduction in the PhoQ dimer. Future studies should be able to test this hypothesis.
Supplementary Material
Acknowledgements
We thank Mark Goulian and Tim Miyashiro for providing the E. coli strain TIM206 and plasmid pTM69, and for helpful discussions. We thank Eric Klein for assistance with Western blotting. This work was supported in part by grants from the NIH (GM54616 and AI74866 to W.F.D., and GM073499 to S.D.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida T, Phadtare S, Inouye M. Functional and Structural Characterization of EnvZ, an Osmosensing Histidine Kinase of E. coli. Methods Enzymol. 2007;423:184–202. doi: 10.1016/S0076-6879(07)23008-3. [DOI] [PubMed] [Google Scholar]
- 4.Gao R, Lynn DG. Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J Bacteriol. 2005;187:2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaspar S, Perozzo R, Reinelt S, Meyer M, Pfister K, Scapozza L, Bott M. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol Microbiol. 1999;33:858–872. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- 8.Garcia Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 9.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 11.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khorchid A, Ikura M. Bacterial histidine kinase as signal sensor and transducer. Int J Biochem Cell Biol. 2006;38:307–312. doi: 10.1016/j.biocel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Milligan DL, Koshland DE., Jr Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 14.Milligan DL, Koshland DE., Jr Purification and characterization of the periplasmic domain of the aspartate chemoreceptor. J Biol Chem. 1993;268:19991–19997. [PubMed] [Google Scholar]
- 15.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 16.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 17.Scott WG, Stoddard BL. Transmembrane signalling and the aspartate receptor. Structure. 1994;2:877–887. doi: 10.1016/s0969-2126(94)00088-3. [DOI] [PubMed] [Google Scholar]
- 18.Khorchid A, Inouye M, Ikura M. Structural characterization of Escherichia coli sensor histidine kinase EnvZ: the periplasmic C-terminal core domain is critical for homodimerization. Biochem J. 2005;385:255–264. doi: 10.1042/BJ20041125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaku H, Mizuno T. The membrane-located osmosensory kinase, EnvZ, that contains a leucine zipper-like motif functions as a dimmer in Escherichia coli. FEBS Lett. 1997;417:409–413. doi: 10.1016/s0014-5793(97)01329-x. [DOI] [PubMed] [Google Scholar]
- 20.Hefti MH, Francoijs KJ, de Vries SC, Dixon R, Vervoort J. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur J Biochem. 2004;271:1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x. [DOI] [PubMed] [Google Scholar]
- 21.Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17:706–715. doi: 10.1016/j.sbi.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 23.Ma X, Sayed N, Baskaran P, Beuve A, van den Akker F. PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J Biol Chem. 2008;283:1167–1178. doi: 10.1074/jbc.M706218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 26.Waldburger CD, Sauer RT. Signal detection by the PhoQ sensor-transmitter. Characterization of the sensor domain and a response-impaired mutant that identifies ligand-binding determinants. J Biol Chem. 1996;271:26630–26636. doi: 10.1074/jbc.271.43.26630. [DOI] [PubMed] [Google Scholar]
- 27.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 28.Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal Structure of a Functional Dimer of the PhoQ Sensor Domain. J Biol Chem. 2008 doi: 10.1074/jbc.M710592200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prost LR, Sanowar S, Miller SI. Salmonella sensing of antimicrobial mechanisms to promote survival within macrophages. Immunol Rev. 2007;219:55–65. doi: 10.1111/j.1600-065X.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 30.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Imer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A Ligand-induced Switch in the Periplasmic Domain of Sensor Histidine Kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 31.Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 32.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 35.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 36.Lai WC, Hazelbauer GL. Analyzing transmembrane chemoreceptors using in vivo disulfide formation between introduced cysteines. Methods Enzymol. 2007;423:299–316. doi: 10.1016/S0076-6879(07)23013-7. [DOI] [PubMed] [Google Scholar]
- 37.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 38.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 39.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai WC, Peach ML, Lybrand TP, Hazelbauer GL. Diagnostic cross-linking of paired cysteine pairs demonstrates homologous structures for two chemoreceptor domains with low sequence identity. Protein Sci. 2006;15:94–101. doi: 10.1110/ps.051802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peach ML, Hazelbauer GL, Lybrand TP. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 2002;11:912–923. doi: 10.1110/ps.4640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dieckmann GR, DeGrado WF. Modeling transmembrane helical oligomers. Curr Opin Struct Biol. 1997;7:486–494. doi: 10.1016/s0959-440x(97)80111-x. [DOI] [PubMed] [Google Scholar]
- 43.Crick FHC. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 44.Xiang Z, Honig B. Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol. 2001;311:421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- 45.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 46.Morona R, Reeves P. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/jb.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.