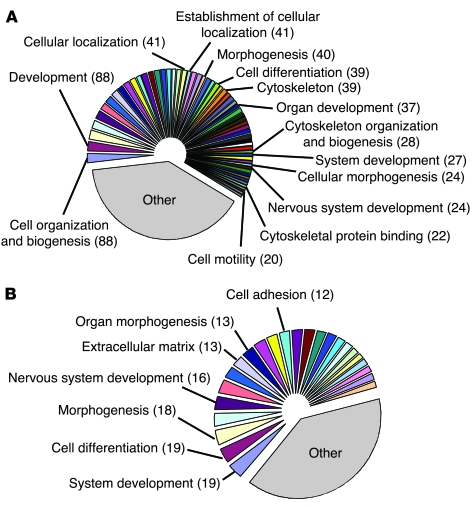
Figure 5. Soluble factors released by myofibroblasts change the gene expression profile of cocultured cholangiocytes.
(A and B) GO analysis of microarray data from cholangiocytes cocultured with MF-HSCs, compared with cholangiocyte monocultures. Each gene probe having an expression ratio above 1.500 and below 0.666 was then assigned to its corresponding GO families. GO sets were therefore ranked based on the total number of genes belonging to them. GO families associated with more than 10% of selected genes were discarded, in order to increase the specificity of the GO analysis. This subpopulation of GO sets was further split into 2 groups, one accounting for at least 60% of this subpopulation, and the other one accounting for the remaining percentage (<40%). A pie chart was then generated using individual names for the first group of GO families, while GO sets belonging to the second group were collectively designated as “other.” (A) Upregulated GO gene sets. (B) Downregulated GO gene sets. Numbers in parenthesis represent altered genes belonging to that specific family. Original pie charts and legends are provided as Supplemental Figures 1 and 2. Complete lists of GO families and EASE scores are reported in Tables 1 and 2 and Supplemental Table 2.